Page 26 - Read Online
P. 26
Page 12 of 16 Lugli et al. Microbiome Res Rep 2023;2:15 https://dx.doi.org/10.20517/mrr.2022.21
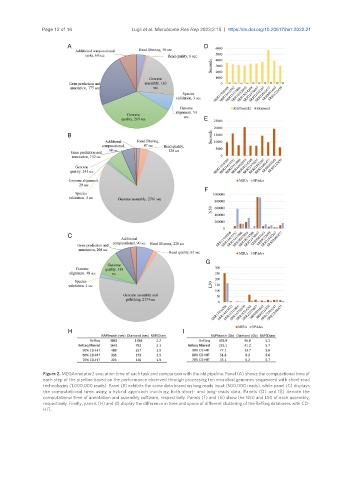
Figure 2. MEGAnnotator2 execution time of each task and comparison with the old pipeline. Panel (A) shows the computational time of
each step of the pipeline based on the performance observed through processing ten microbial genomes sequenced with short-read
technologies (1,000,000 reads). Panel (B) exhibits the same data based on long reads input (500,000 reads), while panel (C) displays
the computational time using a hybrid approach involving both short- and long-reads data. Panels (D) and (E) denote the
computational time of annotation and assembly software, respectively. Panels (F) and (G) show the N50 and L50 of each assembly,
respectively. Finally, panels (H) and (I) display the difference in time and space of different clustering of the RefSeq databases with CD-
HIT.