Page 27 - Read Online
P. 27
Berber et al. J Transl Genet Genom 2021;5:292-303 https://dx.doi.org/10.20517/jtgg.2021.35 Page 298
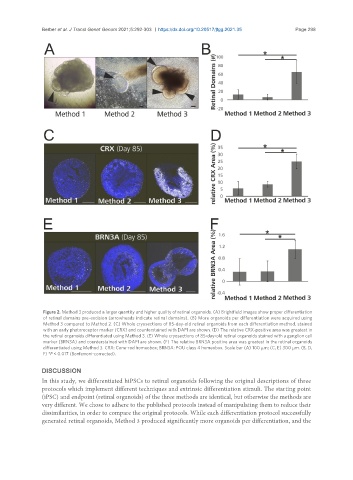
Figure 2. Method 3 produced a larger quantity and higher quality of retinal organoids. (A) Brightfield images show proper differentiation
of retinal domains pre-excision (arrowheads indicate retinal domains). (B) More organoids per differentiation were acquired using
Method 3 compared to Method 2. (C) Whole cryosections of 85-day-old retinal organoids from each differentiation method, stained
with an early photoreceptor marker (CRX) and counterstained with DAPI are shown. (D) The relative CRX-positive area was greatest in
the retinal organoids differentiated using Method 3. (E) Whole cryosections of 85-day-old retinal organoids stained with a ganglion cell
marker (BRN3A) and counterstained with DAPI are shown. (F) The relative BRN3A positive area was greatest in the retinal organoids
differentiated using Method 3. CRX: Cone-rod homeobox; BRN3A: POU class 4 homeobox. Scale bar (A) 100 µm; (C, E) 300 µm. (B, D,
F) *P < 0.017 (Bonferroni-corrected).
DISCUSSION
In this study, we differentiated hiPSCs to retinal organoids following the original descriptions of three
protocols which implement different techniques and extrinsic differentiation stimuli. The starting point
(iPSC) and endpoint (retinal organoids) of the three methods are identical, but otherwise the methods are
very different. We chose to adhere to the published protocols instead of manipulating them to reduce their
dissimilarities, in order to compare the original protocols. While each differentiation protocol successfully
generated retinal organoids, Method 3 produced significantly more organoids per differentiation, and the