Page 90 - Read Online
P. 90
Xu et al. J Transl Genet Genom 2021;5:218-39 https://dx.doi.org/10.20517/jtgg.2021.20 Page 220
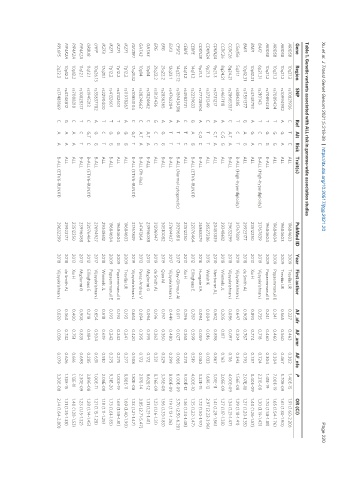
Table 1. Genetic variants associated with ALL risk in genome-wide association studies
Gene Region SNP Ref Alt Risk Trait(s) PubMed ID Year First author AF_afr AF_amr AF_nfe P OR (CI)
ARID5B 10q21.2 rs10821936 C T C ALL 19684603 2009 Treviño LR 0.227 0.463 0.302 1.40E-15 1.91 (1.60-2.20)
ARID5B 10q21.2 rs10994982 A G A ALL 19684603 2009 Treviño LR 0.565 0.560 0.467 5.70E-09 1.61 (1.30-1.90)
ARID5B 10q21.2 rs7089424 T G G ALL 19684604 2009 Papaemmanuil E 0.241 0.460 0.304 7.00E-19 1.65 (1.54-1.76)
ARID5B 10q21.2 rs7089424 T G G B-ALL 19684604 2009 Papaemmanuil E 0.241 0.460 0.304 1.41E-19 1.70 (1.58-1.81)
BAK1 6p21.31 rs210143 T C C B-ALL (High-hyperdiploidy) 31767839 2019 Vijayakrishnan J 0.735 0.718 0.724 2.21E-08 1.30 (1.19-1.43)
BMI1 10p12.31 rs4748793 A G A ALL 23512250 2013 Xu H 0.893 0.774 0.787 8.40E-09 1.40 (1.26-1.57)
BMI1 10p12.31 rs11591377 G A G ALL 29923177 2018 de Smith AJ 0.905 0.767 0.793 2.07E-10 1.27 (1.20-1.35)
C5orf56 5q31.1 rs886285 T C T B-ALL (High-hyperdiploidy) 31767839 2019 Vijayakrishnan J 0.647 0.304 0.343 1.56E-08 1.29 (1.18-1.41)
CCDC26 8q24.21 rs28665337 C A,T A B-ALL 29632299 2018 Vijayakrishnan J 0.086 0.097 0.116 4.00E-09 1.34 (1.21-1.47)
CCDC26 8q24.21 rs4617118 A C,G G ALL 29348612 2018 Wiemels JL 0.305 0.117 0.163 3.05E-09 1.27 (1.17-1.38)
CDKN2A 9p21.3 rs3731217 A C,T A ALL 20453839 2010 Sherborne AL 0.902 0.898 0.867 3.01E-11 1.41 (1.28-1.56)
CDKN2A 9p21.3 rs3731249 C T T ALL 26527286 2015 Walsh K 0.004 0.016 0.033 1.69E-13 2.97 (2.22-3.96)
CDKN2B 9p21.3 rs77728904 A C,G C B-ALL 26868379 2016 Hungate EA 0.094 0.059 0.080 3.32E-15 1.72 (1.50-1.97)
CEBPE 14q11.2 rs2239633 G A G B-ALL (ETV6-RUNX1) 22076464 2012 Ellinghaus E 0.787 0.599 0.519 4.00E-10 1.35 (1.22-1.47)
CEBPE 14q11.2 rs4982731 C T C ALL 23512250 2013 Xu H 0.396 0.368 0.278 9.00E-12 1.36 (1.24-1.48)
CPSF2 14q32.12 rs189434316 A T T B-ALL (Normal cytogenetic) 29296818 2017 Clay-Gilmour AI 0.011 0.027 0.066 6.00E-09 3.70 (2.50-6.20)
ELK3 12q23.1 rs4762284 A T T B-ALL 27694927 2017 Vijayakrishnan J 0.449 0.480 0.299 8.00E-09 1.19 (1.12-1.26)
ERG 21q22.2 rs2836365 A G G B-ALL 30510082 2019 Qian M 0.197 0.360 0.329 3.76E-08 1.56 (1.33-1.83)
ERG 21q22.2 rs8131436 G C C ALL 31296947 2019 de Smith AJ 0.196 0.363 0.331 8.76E-09 1.23 (1.16-1.31)
GATA3 10p14 rs3824662 C A,T A B-ALL 23996088 2013 Migliorini G 0.094 0.395 0.172 8.62E-12 1.31 (1.21-1.41)
GATA3 10p14 rs3824662 C A,T A B-ALL (Ph-like) 24141364 2013 Perez-Andreu V 0.094 0.395 0.172 2.17E-14 3.85 (2.71-5.47)
IGF2BP1 17q21.32 rs10853104 C G,T T B-ALL (ETV6-RUNX1) 31767839 2019 Vijayakrishnan J 0.663 0.420 0.508 1.82E-08 1.33 (1.21-1.47)
IKZF1 7p12.2 rs11978267 A G G ALL 19684603 2009 Treviño LR 0.193 0.241 0.277 8.80E-11 1.69 (1.40-1.90)
IKZF1 7p12.2 rs4132601 T G G ALL 19684604 2009 Papaemmanuil E 0.193 0.242 0.275 1.00E-19 1.69 (1.58-1.81)
IKZF1 7p12.2 rs4132601 T G G B-ALL 19684604 2009 Papaemmanuil E 0.193 0.242 0.275 9.31E-20 1.73 (1.61-1.85)
IKZF3 17q21.1 rs2290400 T C T ALL 29348612 2018 Wiemels JL 0.518 0.619 0.510 2.05E-08 1.18 (1.11-1.25)
LHPP 10q26.13 rs35837782 A G G B-ALL 27694927 2017 Vijayakrishnan J 0.654 0.504 0.631 1.00E-11 1.21 (1.15-1.28)
OR8U8 11q12.1 rs1945213 C G,T C B-ALL (ETV6-RUNX1) 22076464 2012 Ellinghaus E 0.218 0.184 0.285 3.89E-08 1.28 (1.14-1.45)
PIP4K2A 11q12.1 rs10828317 T C T B-ALL 23996088 2013 Migliorini G 0.908 0.835 0.698 2.30E-09 1.23 (1.15-1.32)
PIP4K2A 10p12.2 rs7088318 C A A ALL 23512250 2013 Xu H 0.400 0.751 0.616 1.13E-11 1.40 (1.28-1.53)
PIP4K2A 10p12.2 rs4748812 G A A ALL 29923177 2018 de Smith AJ 0.360 0.742 0.626 1.30E-15 1.31 (1.25-1.38)
RPL6P5 2q22.3 rs17481869 C A A B-ALL (ETV6-RUNX1) 29632299 2018 Vijayakrishnan J 0.020 0.036 0.079 3.20E-08 2.14 (1.64-2.80)