Page 228 - Read Online
P. 228
Page 6 of 11 Zhang et al. J Cancer Metastasis Treat 2020;6:21 I http://dx.doi.org/10.20517/2394-4722.2020.40
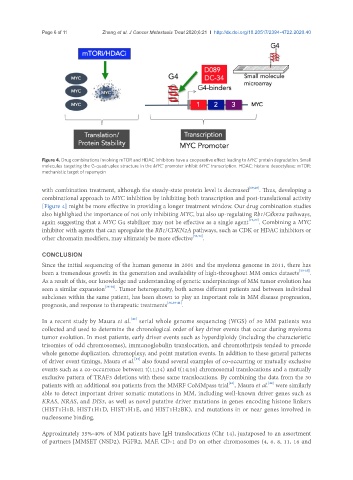
Figure 4. Drug combinations involving mTOR and HDAC inhibitors have a cooperative effect leading to MYC protein degradation. Small
molecules targeting the G-quadruplex structure in the MYC promoter inhibit MYC transcription. HDAC: histone deacetylase; mTOR:
mechanistic target of rapamycin
with combination treatment, although the steady-state protein level is decreased [19,20] . Thus, developing a
combinational approach to MYC inhibition by inhibiting both transcription and post-translational activity
[Figure 4] might be more effective in providing a longer treatment window. Our drug combination studies
also highlighted the importance of not only inhibiting MYC, but also up-regulating Rb1/Cdkn2a pathways,
again suggesting that a MYC G4 stabilizer may not be effective as a single agent [19,20] . Combining a MYC
inhibitor with agents that can upregulate the RB1/CDKN2A pathways, such as CDK or HDAC inhibitors or
other chromatin modifiers, may ultimately be more effective [31,32] .
CONCLUSION
Since the initial sequencing of the human genome in 2001 and the myeloma genome in 2011, there has
been a tremendous growth in the generation and availability of high-throughout MM omics datasets [33-35] .
As a result of this, our knowledge and understanding of genetic underpinnings of MM tumor evolution has
seen a similar expansion [36-38] . Tumor heterogeneity, both across different patients and between individual
subclones within the same patient, has been shown to play an important role in MM disease progression,
prognosis, and response to therapeutic treatments [36,39-41] .
[42]
In a recent study by Maura et al. serial whole genome sequencing (WGS) of 30 MM patients was
collected and used to determine the chronological order of key driver events that occur during myeloma
tumor evolution. In most patients, early driver events such as hyperdiploidy (including the characteristic
trisomies of odd chromosomes), immunoglobulin translocation, and chromothripsis tended to precede
whole genome duplication, chromoplexy, and point mutation events. In addition to these general patterns
[42]
of driver event timings, Maura et al. also found several examples of co-occurring or mutually exclusive
events such as a co-occurrence between t(11;14) and t(14;16) chromosomal translocations and a mutually
exclusive pattern of TRAF3 deletions with these same translocations. By combining the data from the 30
[43]
[42]
patients with an additional 804 patients from the MMRF CoMMpass trial , Maura et al. were similarly
able to detect important driver somatic mutations in MM, including well-known driver genes such as
KRAS, NRAS, and DIS3, as well as novel putative driver mutations in genes encoding histone linkers
(HIST1H1B, HIST1H1D, HIST1H1E, and HIST1H2BK), and mutations in or near genes involved in
nucleosome binding.
Approximately 35%-40% of MM patients have IgH translocations (Chr 14), juxtaposed to an assortment
of partners [MMSET (NSD2), FGFR2, MAF, CD-1 and D3 on other chromosomes (4, 6, 8, 11, 16 and