Page 232 - Read Online
P. 232
Page 6 of 12 Park et al. J Cancer Metastasis Treat 2019;5:17 I http://dx.doi.org/10.20517/2394-4722.2018.84
Table 1. Top 40 network objects ranked by occurrence in top 50 pathway maps
Number of maps Number of maps
Ranking Network objects Ranking Network objects
containing network object containing network object
1 IL-6 24 10 TGF-β receptor type II 8
2 ICAM-1 20 11 NFKBIA 7
3 IL-1 beta 18 11 MHC class II 7
4 p38 MARK 16 12 WNT5A 6
5 VCAM-1 15 12 GRO-2 6
6 VEGF-A 12 12 Tcf (Lef) 6
6 c-Myc 12 12 Bcl-XL 6
7 GM-CSF 11 12 WNT 6
7 CCL2 11 13 GRO-3 5
8 I-kB 10 13 PLAUR 5
8 PTCH1 10 13 IL-1α 5
8 SHH 10 13 SNAIL1 5
9 MMP-1 9 13 PKA-cat 5
10 Fibronectin 8 13 MMP-13 5
10 TCF7L2 8 13 PDGF-R-β 5
10 Frizzled 8 13 CSF1 5
10 TLR2 8 13 PKA-reg 5
10 GLI-2 8 14 Jagged1 4
10 Lef-1 8 14 TRF2 4
Network object ranking reflects the popularity of each network object in pathway map. Smaller rank value means that network object
occurs in greater number of maps. ICAM-1: intercellular adhesion molecule-1
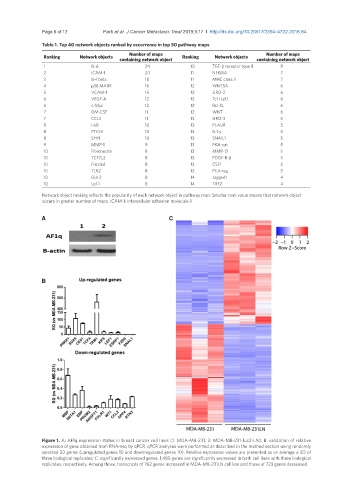
A C
B
Figure 1. A: AF1q expression status in breast cancer cell lines (1: MDA-MB-231, 2: MDA-MB-231-luc2-LN); B: validation of relative
expression of gene obtained from RNA-seq by qPCR. qPCR analyses were performed as described in the method section using randomly
selected 20 genes (upregulated genes 10 and downregulated genes 10). Relative expression values are presented as an average ± SD of
three biological replicates; C: significantly expressed genes. 1,485 genes are significantly expressed in both cell lines with three biological
replicates, respectively. Among those, transcripts of 762 genes increased in MDA-MB-231LN cell line and those of 723 genes decreased