Page 233 - Read Online
P. 233
Park et al. J Cancer Metastasis Treat 2019;5:17 I http://dx.doi.org/10.20517/2394-4722.2018.84 Page 7 of 12
A B
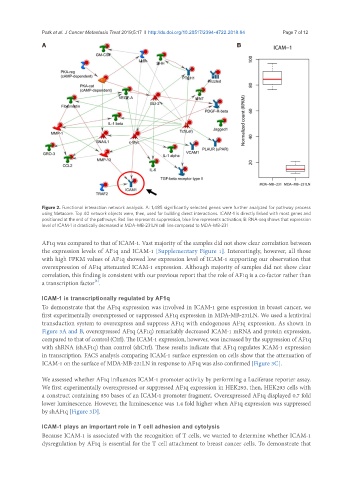
Figure 2. Functional interaction network analysis. A: 1,485 significantly selected genes were further analyzed for pathway process
using Metacore. Top 40 network objects were, then, used for building direct interactions. ICAM-1 is directly linked with most genes and
positioned at the end of the pathways. Red line represents suppression, blue line represents activation; B: RNA-seq shows that expression
level of ICAM-1 is drastically decreased in MDA-MB-231LN cell line compared to MDA-MB-231
AF1q was compared to that of ICAM-1. Vast majority of the samples did not show clear correlation between
the expression levels of AF1q and ICAM-1 [Supplementary Figure 1]. Interestingly, however, all those
with high FPKM values of AF1q showed low expression level of ICAM-1 supporting our observation that
overexpression of AF1q attenuated ICAM-1 expression. Although majority of samples did not show clear
correlation, this finding is consistent with our previous report that the role of AF1q is a co-factor rather than
[2]
a transcription factor .
ICAM-1 is transcriptionally regulated by AF1q
To demonstrate that the AF1q expression was involved in ICAM-1 gene expression in breast cancer, we
first experimentally overexpressed or suppressed AF1q expression in MDA-MB-231LN. We used a lentiviral
transduction system to overexpress and suppress AF1q with endogenous AF1q expression. As shown in
Figure 3A and B, overexpressed AF1q (AF1q) remarkably decreased ICAM-1 mRNA and protein expression,
compared to that of control (Ctrl). The ICAM-1 expression, however, was increased by the suppression of AF1q
with shRNA (shAF1q) than control (shCtrl). These results indicate that AF1q regulates ICAM-1 expression
in transcription. FACS analysis comparing ICAM-1 surface expression on cells show that the attenuation of
ICAM-1 on the surface of MDA-MB-231LN in response to AF1q was also confirmed [Figure 3C].
We assessed whether AF1q influences ICAM-1 promoter activity by performing a Luciferase reporter assay.
We first experimentally overexpressed or suppressed AF1q expression in HEK293, then, HEK293 cells with
a construct containing 850 bases of an ICAM-1 promoter fragment. Overexpressed AF1q displayed 0.7 fold
lower luminescence. However, the luminescence was 1.4 fold higher when AF1q expression was suppressed
by shAF1q [Figure 3D].
ICAM-1 plays an important role in T cell adhesion and cytolysis
Because ICAM-1 is associated with the recognition of T cells, we wanted to determine whether ICAM-1
dysregulation by AF1q is essential for the T cell attachment to breast cancer cells. To demonstrate that