Page 90 - Read Online
P. 90
Page 4 of 9 Othman et al. J Cancer Metastasis Treat 2018;4:50 I http://dx.doi.org/10.20517/2394-4722.2018.41
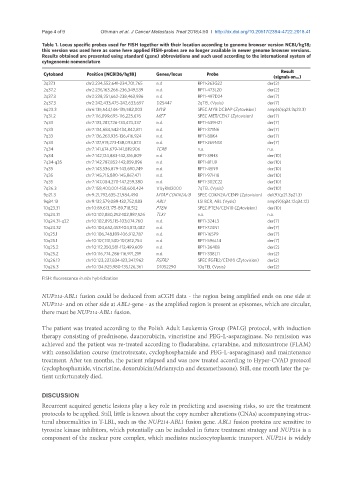
Table 1. Locus specific probes used for FISH together with their location according to genome browser version NCBI/hg18;
this version was used here as some here applied FISH-probes are no longer available in newer genome browser versions.
Results obtained are presented using standard (gene) abbreviations and such used according to the international system of
cytogenomic nomenclature
Result
Cytoband Position [NCBI36/hg18] Genes/locus Probe
(signals on…)
2q37.1 chr2:234,552,641-234,701,765 n.d RP11-263G22 der(2)
2q37.2 chr2:236,163,266-236,349,539 n.d. RP11-473L20 der(2)
2q37.3 chr2:238,251,662-238,463,936 n.d. RP11-497D24 der(7)
2q37.3 chr2:242,433,475-242,633,697 D2S447 2qTEL (Vysis) der(7)
6q23.3 chr6:135,544,146-135,582,003 MYB SPEC MYB DCBAP (Zytovision) amp(6)(q23.3q23.3)
7q31.2 chr7:116,099,695-116,225,676 MET SPEC MET/CEN7 (Zytovision) der(7)
7q33 chr7:133,287,726-133,474,337 n.d. RP11-639H21 der(7)
7q33 chr7:134,684,542-134,842,811 n.d. RP11-371N6 der(7)
7q33 chr7:136,263,935-136,416,924 n.d. RP11-88K4 der(7)
7q33 chr7:137,919,273-138,093,873 n.d. RP11-269N18 der(7)
7q34 chr7:141,674,679-141,819,906 TCRB n.a. n.a.
7q34 chr7:142,124,883-142,316,809 n.d. RP11-39H3 der(10)
7q34-q35 chr7:142,787,852-142,859,896 n.d. RP11-811J9 der(10)
7q35 chr7:143,536,879-143,690,749 n.d. RP11-45N9 der(10)
7q35 chr7:145,715,880-145,867,471 n.d. RP11-97H18 der(10)
7q35 chr7:147,084,270-147,259,380 n.d. RP11-302C22 der(10)
7q36.3 chr7:158,400,001-158,600,424 VIJyRM2000 7qTEL (Vysis) der(10)
9p21.3 chr9:21,792,635-21,984,490 MTAP CDKN2A/B SPEC CDKN2A/CEN9 (Zytovision) del(9)(p21.3p21.3)
9q34.13 chr9:132,579,089-132,752,883 ABL1 LSI BCR, ABL (Vysis) amp(9)(q34.12q34.12)
10q23.31 chr10:89,613,175-89,718,512 PTEN SPEC PTEN/CEN10 (Zytovision) der(10)
10q24.31 chr10:102,880,252-102,887,526 TLX1 n.a. n.a.
10q24.31-q32 chr10:102,895,115-103,074,760 n.d. RP11-324L3 der(7)
10q24.32 chr10:104,652,453-104,813,482 n.d. RP11-724N1 der(7)
10q25.1 chr10:106,748,189-106,912,787 n.d. RP11-165P9 der(7)
10q25.1 chr10:107,741,530-107,812,754 n.d. RP11-596L14 der(7)
10q25.2 chr10:112,350,581-112,499,609 n.d. RP11-364E8 der(2)
10q25.2 chr10:116,774,286-116,971,219 n.d. RP11-338L11 der(2)
10q26.13 chr10:123,227,834-123,347,962 FGFR2 SPEC FGFR2/CEN10 (Zytovision) der(2)
10q26.3 chr10:134,925,980-135,126,361 D10S2290 10qTEL (Vysis) der(2)
FISH: fluorescence in situ hybridization
NUP214-ABL1 fusion could be deduced from aCGH data - the region being amplified ends on one side at
NUP214- and on other side at ABL1-gene - as the amplified region is present as episomes, which are circular,
there must be NUP214-ABL1 fusion.
The patient was treated according to the Polish Adult Leukemia Group (PALG) protocol, with induction
therapy consisting of prednisone, daunorubicin, vincristine and PEG-L-asparaginase. No remission was
achieved and the patient was re-treated according to fludarabine, cytarabine, and mitoxantrone (FLAM)
with consolidation course (metrotrexate, cyclophosphamide and PEG-L-asparaginase) and maintenance
treatment. After ten months, the patient relapsed and was now treated according to Hyper-CVAD protocol
(cyclophosphamide, vincristine, doxorubicin/Adriamycin and dexamethasone). Still, one month later the pa-
tient unfortunately died.
DISCUSSION
Recurrent acquired genetic lesions play a key role in predicting and assessing risks, so are the treatment
protocols to be applied. Still, little is known about the copy number alterations (CNAs) accompanying struc-
tural abnormalities in T-LBL, such as the NUP214-ABL1 fusion gene. ABL1 fusion proteins are sensitive to
tyrosine kinase inhibitors, which potentially can be included in future treatment strategy and NUP214 is a
component of the nuclear pore complex, which mediates nucleocytoplasmic transport. NUP214 is widely