Page 14 - Read Online
P. 14
Page 8 of 14 Gottlieb et al. J Cancer Metastasis Treat 2018;4:37 I http://dx.doi.org/10.20517/2394-4722.2018.26
Exon 7 Number of reads
19,248 7104 3729 1260 6188 2993
Number of mutants
AAA AAT CAA
824 2471A AA (ins A) Asn Gln Lys fs 5
AAA
AAA AAT CAA
825 2472T TA (ins A) Gln fs 21
AAA
AAA AAT CAA
825 2473C CA (ins A) Gln Lys Gln Lys fs 4
AAA
Exon 8 Number of reads
3443 3836 4430 1569 1662 1795
Number of mutants
880 2638T TT (ins T) ACT TTT GAC Asp Stop 4
887 2661G A ATG GTG Met Ile 2 PCa
890 2670G A GTG CAC Val Val 2 2
893 2678C T TTT CCG GAA Pro Leu 2 CAIS
893 2678C CC (ins C) TTT CCG GAA Pro Pro fs 2
893 2679G A TTT CCG GAA Pro Pro 2
905 2715C CC (ins C) GTG CCC AAG Pro Pro fs 3
n/a: not available; PCa: prostate cancer; CAIS: complete androgen insensitivity syndrome
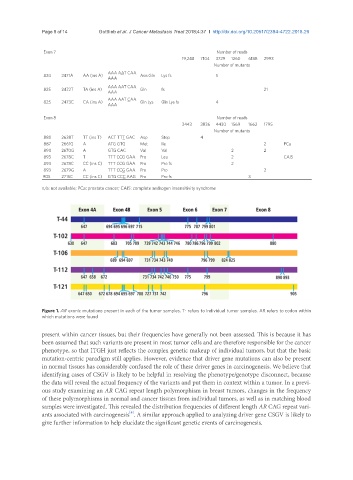
Figure 1. AR exonic mutations present in each of the tumor samples. T- refers to individual tumor samples. AR refers to codon within
which mutations were found
present within cancer tissues, but their frequencies have generally not been assessed. This is because it has
been assumed that such variants are present in most tumor cells and are therefore responsible for the cancer
phenotype, so that ITGH just reflects the complex genetic makeup of individual tumors, but that the basic
mutation-centric paradigm still applies. However, evidence that driver gene mutations can also be present
in normal tissues has considerably confused the role of these driver genes in carcinogenesis. We believe that
identifying cases of CSGV is likely to be helpful in resolving the phenotype/genotype disconnect, because
the data will reveal the actual frequency of the variants and put them in context within a tumor. In a previ-
ous study examining an AR CAG repeat length polymorphism in breast tumors, changes in the frequency
of these polymorphisms in normal and cancer tissues from individual tumors, as well as in matching blood
samples were investigated. This revealed the distribution frequencies of different length AR CAG repeat vari-
[6]
ants associated with carcinogenesis . A similar approach applied to analyzing driver gene CSGV is likely to
give further information to help elucidate the significant genetic events of carcinogenesis.