Page 53 - Read Online
P. 53
Page 6 of 11 Chang et al. J Cancer Metastasis Treat 2019;5:78 I http://dx.doi.org/10.20517/2394-4722.2019.31
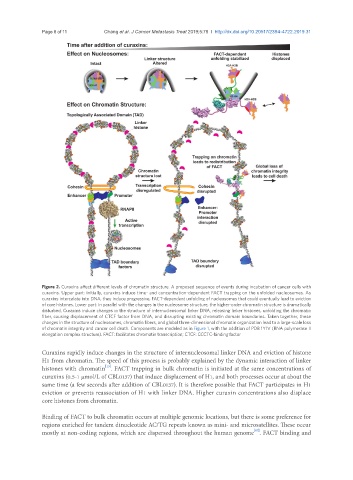
Figure 2. Curaxins affect different levels of chromatin structure. A proposed sequence of events during incubation of cancer cells with
curaxins. Upper part: initially, curaxins induce time- and concentration-dependent FACT trapping on the unfolded nucleosomes. As
curaxins intercalate into DNA, they induce progressive, FACT-dependent unfolding of nucleosomes that could eventually lead to eviction
of core histones. Lower part: in parallel with the changes in the nucleosome structure, the higher-order chromatin structure is dramatically
disturbed. Curaxins induce changes in the structure of internucleosomal linker DNA, releasing linker histones, unfolding the chromatin
fiber, causing displacement of CTCF factor from DNA, and disrupting existing chromatin domain boundaries. Taken together, these
changes in the structure of nucleosomes, chromatin fibers, and global three-dimensional chromatin organization lead to a large-scale loss
of chromatin integrity and cancer cell death. Components are modeled as in Figure 1, with the addition of PDB 1Y1V (RNA polymerase II
elongation complex structure). FACT: facilitates chromatin transcription; CTCF: CCCTC-binding factor
Curaxins rapidly induce changes in the structure of internucleosomal linker DNA and eviction of histone
H1 from chromatin. The speed of this process is probably explained by the dynamic interaction of linker
[13]
histones with chromatin . FACT trapping in bulk chromatin is initiated at the same concentrations of
curaxins (0.5-1 mmol/L of CBL0137) that induce displacement of H1, and both processes occur at about the
same time (a few seconds after addition of CBL0137). It is therefore possible that FACT participates in H1
eviction or prevents reassociation of H1 with linker DNA. Higher curaxin concentrations also displace
core histones from chromatin.
Binding of FACT to bulk chromatin occurs at multiple genomic locations, but there is some preference for
regions enriched for tandem dinucleotide AC/TG repeats known as mini- and microsatellites. These occur
[87]
mostly at non-coding regions, which are dispersed throughout the human genome . FACT binding and