Page 108 - Read Online
P. 108
Page 206 Tutanov et al. Extracell Vesicles Circ Nucleic Acids 2023;4:195-217 https://dx.doi.org/10.20517/evcna.2023.17
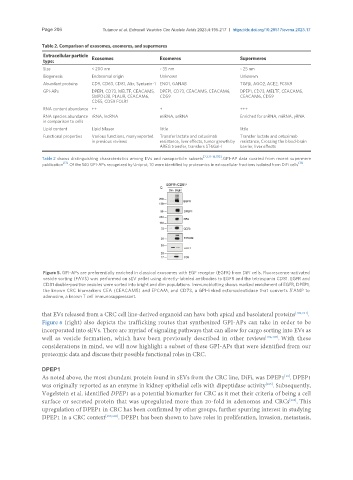
Table 2. Comparison of exosomes, exomeres, and supermeres
Extracellular particle Exosomes Exomeres Supermeres
type:
Size < 200 nm ~ 35 nm ~ 25 nm
Biogenesis Endosomal origin Unknown Unknown
Abundant proteins CD9, CD63, CD81, Alix, Syntenin-1 ENO1, GANAB TGFβi, AGO2, ACE2, PCSK9
GPI-APs DPEP1, CD73, MELTF, CEACAM5, DPEP1, CD73, CEACAM5, CEACAM6, DPEP1, CD73, MELTF, CEACAM5,
SMPDL3B, PLAUR, CEACAM6, CD59 CEACAM6, CD59
CD55, CD59 FOLR1
RNA content abundance ++ + +++
RNA species abundance rRNA, lncRNA miRNA, snRNA Enriched for snRNA, miRNA, yRNA
in comparison to cells
Lipid content Lipid bilayer little little
Functional properties Various functions, many reported Transfer lactate and cetuximab Transfer lactate and cetuximab
in previous reviews resistance, liver effects, tumor growth by resistance, Crossing the blood-brain
AREG transfer, transfers ST6Gal-I barrier, liver effects
[2,3,12-14,170]
Table 2 shows distinguishing characteristics among EVs and nanoparticle subsets . GPI-AP data curated from recent supermere
publication [13] . Of the 140 GPI-APs recognized by Uniprot, 10 were identified by proteomics in extracellular fractions isolated from DiFi cells [13] .
Figure 5. GPI-APs are preferentially enriched in classical exosomes with EGF receptor (EGFR) from DiFi cells. Fluorescence-activated
vesicle sorting (FAVS) was performed on sEV pellet using directly-labeled antibodies to EGFR and the tetraspanin CD81. EGFR and
CD81 double-positive vesicles were sorted into bright and dim populations. Immunoblotting shows marked enrichment of EGFR, DPEP1,
the known CRC biomarkers CEA (CEACAM5) and EPCAM, and CD73, a GPI-linked ectonucleotidase that converts 5’AMP to
adenosine, a known T cell immunosuppressant.
that EVs released from a CRC cell line-derived organoid can have both apical and basolateral proteins [100,101] .
Figure 6 (right) also depicts the trafficking routes that synthesized GPI-APs can take in order to be
incorporated into sEVs. There are myriad of signaling pathways that can allow for cargo sorting into EVs as
well as vesicle formation, which have been previously described in other reviews [102,103] . With these
considerations in mind, we will now highlight a subset of these GPI-APs that were identified from our
proteomic data and discuss their possible functional roles in CRC.
DPEP1
[13]
As noted above, the most abundant protein found in sEVs from the CRC line, DiFi, was DPEP1 . DPEP1
was originally reported as an enzyme in kidney epithelial cells with dipeptidase activity . Subsequently,
[105]
Vogelstein et al. identified DPEP1 as a potential biomarker for CRC as it met their criteria of being a cell
[106]
surface or secreted protein that was upregulated more than 20-fold in adenomas and CRCs . This
upregulation of DPEP1 in CRC has been confirmed by other groups, further spurring interest in studying
DPEP1 in a CRC context [107,108] . DPEP1 has been shown to have roles in proliferation, invasion, metastasis,