Page 123 - Read Online
P. 123
Zhong et al. Chem Synth 2023;3:27 https://dx.doi.org/10.20517/cs.2023.15 Page 17 of 25
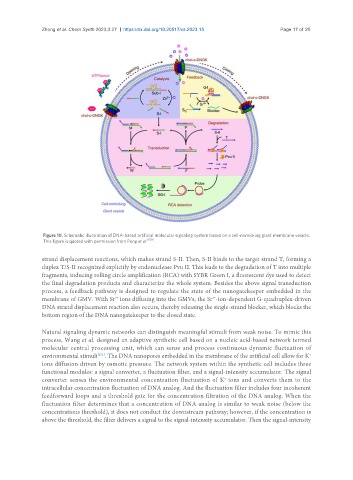
Figure 10. Schematic illustration of DNA-based artificial molecular signaling system based on a cell-mimicking giant membrane vesicle.
This figure is quoted with permission from Peng et al. [150] .
strand displacement reactions, which makes strand S-II. Then, S-II binds to the target strand T, forming a
duplex T/S-II recognized explicitly by endonuclease Pvu II. This leads to the degradation of T into multiple
fragments, inducing rolling circle amplification (RCA) with SYBR Green I, a fluorescent dye used to detect
the final degradation products and characterize the whole system. Besides the above signal transduction
process, a feedback pathway is designed to regulate the state of the nanogatekeeper embedded in the
2+
2+
membrane of GMV. With Sr ions diffusing into the GMVs, the Sr -ion-dependent G-quadruplex-driven
DNA strand displacement reaction also occurs, thereby releasing the single-strand blocker, which blocks the
bottom region of the DNA nanogatekeeper to the closed state.
Natural signaling dynamic networks can distinguish meaningful stimuli from weak noise. To mimic this
process, Wang et al. designed an adaptive synthetic cell based on a nucleic acid-based network termed
molecular central processing unit, which can sense and process continuous dynamic fluctuation of
environmental stimuli . The DNA nanopores embedded in the membrane of the artificial cell allow for K
+
[151]
ions diffusion driven by osmotic pressure. The network system within the synthetic cell includes three
functional modules: a signal converter, a fluctuation filter, and a signal-intensity accumulator. The signal
converter senses the environmental concentration fluctuation of K ions and converts them to the
+
intracellular concentration fluctuation of DNA analog. And the fluctuation filter includes four incoherent
feedforward loops and a threshold gate for the concentration filtration of the DNA analog. When the
fluctuation filter determines that a concentration of DNA analog is similar to weak noise (below the
concentrations threshold), it does not conduct the downstream pathway; however, if the concentration is
above the threshold, the filter delivers a signal to the signal-intensity accumulator. Then the signal-intensity