Page 122 - Read Online
P. 122
Page 16 of 25 Zhong et al. Chem Synth 2023;3:27 https://dx.doi.org/10.20517/cs.2023.15
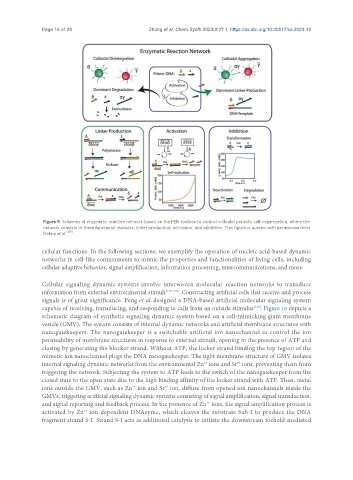
Figure 9. Schemes of enzymatic reaction network based on the PEN toolbox to control colloidal periodic self-organization, where the
network consists of three functional modules, linker production, activation, and inhibition. This figure is quoted with permission from
Dehne et al. [139] .
cellular functions. In the following sections, we exemplify the operation of nucleic acid-based dynamic
networks in cell-like containments to mimic the properties and functionalities of living cells, including
cellular adaptive behavior, signal amplification, information processing, intercommunications, and more.
Cellular signaling dynamic systems involve interwoven molecular reaction networks to transduce
information from external environmental stimuli [148-149] . Constructing artificial cells that receive and process
signals is of great significance. Peng et al. designed a DNA-based artificial molecular signaling system
capable of receiving, transducing, and responding to calls from an outside stimulus . Figure 10 depicts a
[150]
schematic diagram of synthetic signaling dynamic system based on a cell-mimicking giant membrane
vesicle (GMV). The system consists of internal dynamic networks and artificial membrane structures with
nanogatekeepers. The nanogatekeeper is a switchable artificial ion nanochannel to control the ion
permeability of membrane structures in response to external stimuli, opening in the presence of ATP and
closing by generating the blocker strand. Without ATP, the locker strand binding the top region of the
mimetic ion nanochannel plugs the DNA nanogatekeeper. The tight membrane structure of GMV isolates
internal signaling dynamic networks from the environmental Zn ions and Sr ions, preventing them from
2+
2+
triggering the network. Subjecting the system to ATP leads to the switch of the nanogatekeeper from the
closed state to the open state due to the high binding affinity of the locker strand with ATP. Then, metal
2+
2+
ions outside the GMV, such as Zn ion and Sr ion, diffuse from opened ion nanochannels inside the
GMVs, triggering artificial signaling dynamic systems consisting of signal amplification, signal transduction,
and signal reporting and feedback process. In the presence of Zn ions, the signal amplification process is
2+
activated by Zn -ion-dependent DNAzyme, which cleaves the substrate Sub-I to produce the DNA
2+
fragment strand S-I. Strand S-I acts as additional catalysis to initiate the downstream toehold-mediated