Page 120 - Read Online
P. 120
Page 14 of 25 Zhong et al. Chem Synth 2023;3:27 https://dx.doi.org/10.20517/cs.2023.15
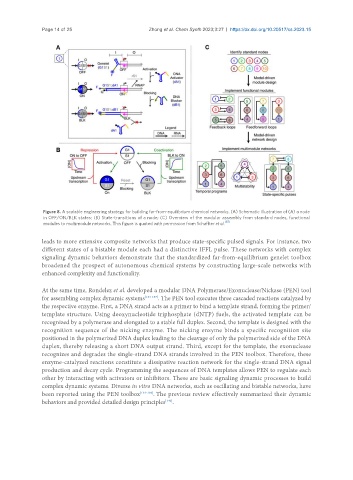
Figure 8. A scalable engineering strategy for building far-from-equilibrium chemical networks. (A) Schematic illustration of (A) a node
in OFF/ON/BLK states; (B) State-transitions of a node; (C) Overview of the modular assembly from standard nodes, functional
modules to multimodule networks. This figure is quoted with permission from Schaffter et al. [87]
leads to more extensive composite networks that produce state-specific pulsed signals. For instance, two
different states of a bistable module each had a distinctive IFFL pulse. These networks with complex
signaling dynamic behaviors demonstrate that the standardized far-from-equilibrium genelet toolbox
broadened the prospect of autonomous chemical systems by constructing large-scale networks with
enhanced complexity and functionality.
At the same time, Rondelez et al. developed a modular DNA Polymerase/Exonuclease/Nickase (PEN) tool
for assembling complex dynamic systems [134-137] . The PEN tool executes three cascaded reactions catalyzed by
the respective enzyme. First, a DNA strand acts as a primer to bind a template strand, forming the primer/
template structure. Using deoxynucleotide triphosphate (dNTP) fuels, the activated template can be
recognized by a polymerase and elongated to a stable full duplex. Second, the template is designed with the
recognition sequence of the nicking enzyme. The nicking enzyme binds a specific recognition site
positioned in the polymerized DNA duplex leading to the cleavage of only the polymerized side of the DNA
duplex, thereby releasing a short DNA output strand. Third, except for the template, the exonuclease
recognizes and degrades the single-strand DNA strands involved in the PEN toolbox. Therefore, these
enzyme-catalyzed reactions constitute a dissipative reaction network for the single-strand DNA signal
production and decay cycle. Programming the sequences of DNA templates allows PEN to regulate each
other by interacting with activators or inhibitors. These are basic signaling dynamic processes to build
complex dynamic systems. Diverse in vitro DNA networks, such as oscillating and bistable networks, have
been reported using the PEN toolbox [134-139] . The previous review effectively summarized their dynamic
[140]
behaviors and provided detailed design principles .