Page 117 - Read Online
P. 117
Zhong et al. Chem Synth 2023;3:27 https://dx.doi.org/10.20517/cs.2023.15 Page 11 of 25
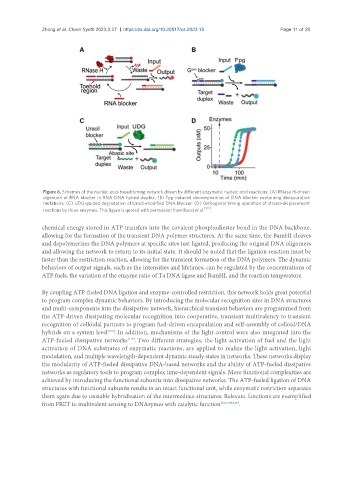
Figure 6. Schemes of the nucleic acid-based timing network driven by different enzymatic nucleic acid reactions: (A) RNase H-driven
digestion of RNA blocker in RNA-DNA hybrid duplex; (B) Fpg-induced decomposition of DNA blocker containing deoxyuridine
mutations; (C) UDG-guided degradation of Uracil-modified DNA blocker. (D) Orthogonal timing operation of strand-displacement
reactions by three enzymes. This figure is quoted with permission from Bucci et al. [122] .
chemical energy stored in ATP transfers into the covalent phosphodiester bond in the DNA backbone,
allowing for the formation of the transient DNA polymer structures. At the same time, the BamHI cleaves
and depolymerizes the DNA polymers at specific sites just ligated, producing the original DNA oligomers
and allowing the network to return to its initial state. It should be noted that the ligation reaction must be
faster than the restriction reaction, allowing for the transient formation of the DNA polymers. The dynamic
behaviors of output signals, such as the intensities and lifetimes, can be regulated by the concentrations of
ATP fuels, the variation of the enzyme ratio of T4 DNA ligase and BamHI, and the reaction temperature.
By coupling ATP-fueled DNA ligation and enzyme-controlled restriction, this network holds great potential
to program complex dynamic behaviors. By introducing the molecular recognition sites in DNA structures
and multi-components into the dissipative network, hierarchical transient behaviors are programmed from
the ATP-driven dissipating molecular recognition into cooperative, transient multivalency to transient
recognition of colloidal partners to program fuel-driven encapsulation and self-assembly of colloid/DNA
hybrids on a system level . In addition, mechanisms of the light control were also integrated into the
[109]
ATP-fueled dissipative networks . Two different strategies, the light activation of fuel and the light
[115]
activation of DNA substrates of enzymatic reactions, are applied to realize the light activation, light
modulation, and multiple wavelength-dependent dynamic steady states in networks. These networks display
the modularity of ATP-fueled dissipative DNA-based networks and the ability of ATP-fueled dissipative
networks as regulatory tools to program complex time-dependent signals. More functional complexities are
achieved by introducing the functional subunits into dissipative networks. The ATP-fueled ligation of DNA
structures with functional subunits results in an intact functional unit, while enzymatic restriction separates
them again due to unstable hybridization of the intermediate structures. Relevant functions are exemplified
from FRET to multivalent sensing to DNAzymes with catalytic function [107,109,126] .