Page 65 - Read Online
P. 65
Kimbowa et al. Art Int Surg 2024;4:149-69 https://dx.doi.org/10.20517/ais.2024.20 Page 159
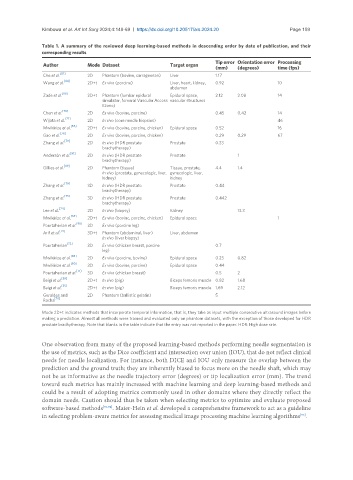
Table 1. A summary of the reviewed deep learning-based methods in descending order by date of publication, and their
corresponding results
Tip error Orientation error Processing
Author Mode Dataset Target organ
(mm) (degrees) time (fps)
[87]
Che et al. 2D Phantom (bovine, carrageenan) Liver 1.17
Wang et al. [84] 2D+t Ex vivo (porcine) Liver, heart, kidney, 0.92 10
abdomen
[88]
Zade et al. 2D+t Phantom (lumbar epidural Epidural space, 2.12 2.08 14
simulator, femoral Vascular Access vascular structures
Ezono)
Chen et al. [78] 2D Ex vivo (bovine, porcine) 0.45 0.42 14
[77]
Wijata et al. 2D In vivo (core needle biopsies) 46
Mwikirize et al. [66] 2D+t Ex vivo (bovine, porcine, chicken) Epidural space 0.52 16
[76]
Gao et al. 2D Ex vivo (bovine, porcine, chicken) 0.29 0.29 67
Zhang et al. [79] 2D In vivo (HDR prostate Prostate 0.33
brachytherapy)
Andersén et al. [81] 2D In vivo (HDR prostate Prostate 1
brachytherapy)
[89]
Gillies et al. 2D Phantom (tissue) Tissue, prostate, 4.4 1.4
In vivo (prostate, gynecologic, liver, gynecologic, liver,
kidney) kidney
Zhang et al. [79] 3D In vivo (HDR prostate Prostate 0.44
brachytherapy)
Zhang et al. [75] 3D In vivo (HDR prostate Prostate 0.442
brachytherapy)
Lee et al. [74] 2D In vivo (biopsy) Kidney 13.3
[65]
Mwikirize et al. 2D+t Ex vivo (bovine, porcine, chicken) Epidural space 1
Pourtaherian et al. [90] 3D Ex vivo (porcine leg)
[73]
Arif et al. 3D+t Phantom (abdominal, liver) Liver, abdomen
In vivo (liver biopsy)
[72]
Pourtaherian 3D Ex vivo (chicken breast, porcine 0.7
leg)
Mwikirize et al. [83] 2D Ex vivo (porcine, bovine) Epidural space 0.23 0.82
[60]
Mwikirize et al. 3D Ex vivo (bovine, porcine) Epidural space 0.44
Pourtaherian et al. [71] 3D Ex vivo (chicken breast) 0.5 2
[69]
Beigi et al. 2D+t In vivo (pig) Biceps femoris muscle 0.82 1.68
Beigi et al. [55] 2D+t In vivo (pig) Biceps femoris muscle 1.69 2.12
Geraldes and 2D Phantom (ballistic gelatin) 5
Rocha [70]
Mode 2D+t indicates methods that incorporate temporal information, that is, they take as input multiple consecutive ultrasound images before
making a prediction. Almost all methods were trained and evaluated only on phantom datasets, with the exception of those developed for HDR
prostate brachytherapy. Note that blanks in the table indicate that the entry was not reported in the paper. HDR: High dose rate.
One observation from many of the proposed learning-based methods performing needle segmentation is
the use of metrics, such as the Dice coefficient and intersection over union (IOU), that do not reflect clinical
needs for needle localization. For instance, both DICE and IOU only measure the overlap between the
prediction and the ground truth; they are inherently biased to focus more on the needle shaft, which may
not be as informative as the needle trajectory error (degrees) or tip localization error (mm). The trend
toward such metrics has mainly increased with machine learning and deep learning-based methods and
could be a result of adopting metrics commonly used in other domains where they directly reflect the
domain needs. Caution should thus be taken when selecting metrics to optimize and evaluate proposed
software-based methods [93,94] . Maier-Hein et al. developed a comprehensive framework to act as a guideline
[93]
in selecting problem-aware metrics for assessing medical image processing machine learning algorithms .