Page 145 - Read Online
P. 145
Page 6 of 10 Sobenin et al. Vessel Plus 2019;3:14 I http://dx.doi.org/10.20517/2574-1209.2018.63
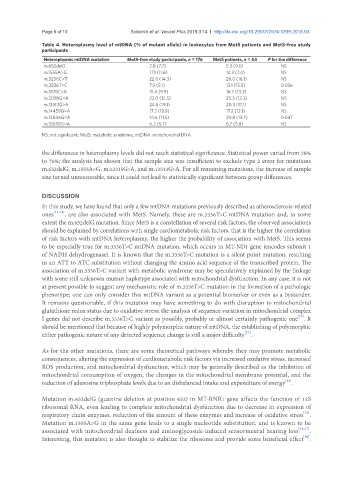
Table 4. Heteroplasmy level of mtDNA (% of mutant allele) in leukocytes from MetS patients and MetS-free study
participants
Heteroplasmic mtDNA mutation MetS-free study participants, n = 176 MetS patients, n = 44 P for the difference
m.652delG 2.8 (7.7) 5.3 (9.0) NS
m.1555A>G 17.1 (11.6) 14.3 (7.4) NS
m.3256C>T 22.6 (14.3) 26.0 (16.1) NS
m.3336T>C 7.9 (5.1) 13.1 (15.5) 0.036
m.5178C>A 15.4 (9.9) 16.7 (13.3) NS
m.12315G>A 22.0 (12.5) 25.3 (12.3) NS
m.13513G>A 24.6 (19.1) 20.3 (17.1) NS
m.14459G>A 17.2 (12.9) 17.2 (12.1) NS
m.14846G>A 14.6 (11.5) 20.8 (13.7) 0.047
m.15059G>A 6.2 (5.7) 6.7 (5.8) NS
NS: not significant; MetS: metabolic syndrome; mtDNA: mitochondrial DNA
the differences in heteroplasmy levels did not reach statistical significance. Statistical power varied from 28%
to 76%; the analysis has shown that the sample size was insufficient to exclude type 2 error for mutations
m.652delG, m.1555A>G, m.12315G>A, and m.13513G>A. For all remaining mutations, the increase of sample
size turned unreasonable, since it could not lead to statistically significant between-group differences.
DISCUSSION
In this study, we have found that only a few mtDNA mutations previously described as atherosclerosis-related
ones [13-18] , are also associated with MetS. Namely, these are m.3336T>C mtDNA mutation and, to some
extent the m.652delG mutation. Since MetS is a constellation of several risk factors, the observed associations
should be explained by correlations with single cardiometabolic risk factors, that is the higher the correlation
of risk factors with mtDNA heteroplasmy, the higher the probability of association with MetS. This seems
to be especially true for m.3336T>C mtDNA mutation, which occurs in MT-ND1 gene (encodes subunit 1
of NADH dehydrogenase). It is known that the m.3336T>C mutation is a silent point mutation, resulting
in an ATT to ATC substitution without changing the amino acid sequence of the transcribed protein. The
association of m.3336T>C variant with metabolic syndrome may be speculatively explained by the linkage
with some still unknown mutant haplotype associated with mitochondrial dysfunction. In any case, it is not
at present possible to suggest any mechanistic role of m.3336T>C mutation in the formation of a pathologic
phenotype; one can only consider this mtDNA variant as a potential biomarker or even as a bystander.
It remains questionable, if this mutation may have something to do with disruption to mitochondrial
glutathione redox status due to oxidative stress; the analysis of sequence variation in mitochondrial complex
[23]
I genes did not describe m.3336T>C variant as possibly, probably or almost certainly pathogenic one . It
should be mentioned that because of highly polymorphic nature of mtDNA, the establishing of polymorphic
[23]
either pathogenic nature of any detected sequence change is still a major difficulty .
As for the other mutations, there are some theoretical pathways whereby they may promote metabolic
consequences, altering the expression of cardiometabolic risk factors via increased oxidative stress, increased
ROS production, and mitochondrial dysfunction, which may be generally described as the inhibition of
mitochondrial consumption of oxygen, the changes in the mitochondrial membrane potential, and the
[24]
reduction of adenosine triphosphate levels due to an disbalanced intake and expenditure of energy .
Mutation m.652delG (guanine deletion at position 652) in MT-RNR1 gene affects the function of 12S
ribosomal RNA, even leading to complete mitochondrial dysfunction due to decrease in expression of
[25]
respiratory chain enzymes, reduction of the amount of these enzymes and increase of oxidative stress .
Mutation m.1555A>G in the same gene leads to a single nucleotide substitution, and is known to be
associated with mitochondrial deafness and aminoglycoside-induced sensorineural hearing loss [26,27] .
[20]
Interesting, this mutation is also thought to stabilize the ribosome and provide some beneficial effect .