Page 150 - Read Online
P. 150
Maiocchi et al. Vessel Plus 2023;7:27 https://dx.doi.org/10.20517/2574-1209.2023.69 Page 13 of 19
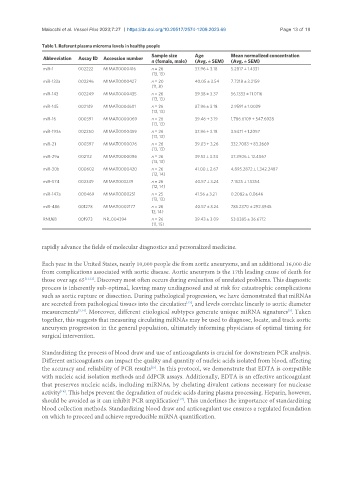
Table 1. Referent plasma microrna levels in healthy people
Sample size Age Mean normalized concentration
Abbreviation Assay ID Accession number
n (female, male) (Avg. ± SEM) (Avg. ± SEM)
miR-1 002222 MIMAT0000416 n = 26 37.96 ± 3.18 5.2817 ± 1.4331
(13, 13)
miR-133a 002246 MIMAT0000427 n = 20 40.05 ± 3.54 7.7218 ± 3.2159
(11, 8)
miR-143 002249 MIMAT0000435 n = 26 39.38 ± 3.37 56.1333 ± 11.0116
(13, 13)
miR-145 002149 MIMAT0004601 n = 26 37.96 ± 3.18 2.9591 ± 1.0029
(13, 13)
miR-16 000391 MIMAT0000069 n = 26 39.46 ± 3.19 1,786.6109 ± 547.6928
(13, 13)
miR-193a 002250 MIMAT0000459 n = 26 37.96 ± 3.18 3.5471 ± 1.2057
(13, 13)
miR-21 000397 MIMAT0000076 n = 26 39.03 ± 3.26 332.7083 ± 83.2669
(13, 13)
miR-29a 002112 MIMAT0000086 n = 26 39.53 ± 3.34 37.3906 ± 12.4067
(13, 13)
miR-30b 000602 MIMAT0000420 n = 26 41.00 ± 2.67 4,895.2872 ± 1,342.2487
(12, 14)
miR-574 002349 MIMAT003239 n = 26 40.57 ± 3.24 7.1525 ± 1.5354
(12, 14)
miR-147a 000469 MIMAT0000251 n = 25 41.56 ± 3.21 0.2082 ± 0.0646
(12, 13)
miR-486 001278 MIMAT0002177 n = 26 40.57 ± 3.24 785.2370 ± 292.3945
12, 14)
RNU6B 001973 NR_004394 n = 26 39.43 ± 3.09 53.0385 ± 26.6712
(11, 15)
rapidly advance the fields of molecular diagnostics and personalized medicine.
Each year in the United States, nearly 10,000 people die from aortic aneurysms, and an additional 16,000 die
from complications associated with aortic disease. Aortic aneurysm is the 17th leading cause of death for
those over age 65 [11,12] . Discovery most often occurs during evaluation of unrelated problems. This diagnostic
process is inherently sub-optimal, leaving many undiagnosed and at risk for catastrophic complications
such as aortic rupture or dissection. During pathological progression, we have demonstrated that miRNAs
[13]
are secreted from pathological tissues into the circulation , and levels correlate linearly to aortic diameter
measurements [3,14] . Moreover, different etiological subtypes generate unique miRNA signatures . Taken
[3]
together, this suggests that measuring circulating miRNAs may be used to diagnose, locate, and track aortic
aneurysm progression in the general population, ultimately informing physicians of optimal timing for
surgical intervention.
Standardizing the process of blood draw and use of anticoagulants is crucial for downstream PCR analysis.
Different anticoagulants can impact the quality and quantity of nucleic acids isolated from blood, affecting
the accuracy and reliability of PCR results . In this protocol, we demonstrate that EDTA is compatible
[15]
with nucleic acid isolation methods and ddPCR assays. Additionally, EDTA is an effective anticoagulant
that preserves nucleic acids, including miRNAs, by chelating divalent cations necessary for nuclease
activity . This helps prevent the degradation of nucleic acids during plasma processing. Heparin, however,
[16]
[17]
should be avoided as it can inhibit PCR amplification . This underlines the importance of standardizing
blood collection methods. Standardizing blood draw and anticoagulant use ensures a regulated foundation
on which to proceed and achieve reproducible miRNA quantification.