Page 9 - Read Online
P. 9
Shetty et al. Microbiome Res Rep 2023;2:14 https://dx.doi.org/10.20517/mrr.2022.18 Page 3 of 5
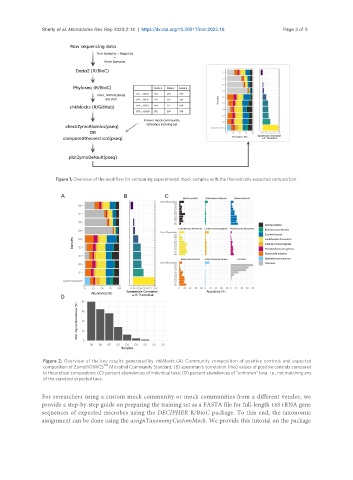
Figure 1. Overview of the workflow for comparing experimental mock samples with the theoretically expected composition.
Figure 2. Overview of the key results generated by chkMocks. (A) Community composition of positive controls and expected
TM
composition of ZymoBIOMICS Microbial Community Standard; (B) spearman’s correlation (rho) values of positive controls compared
to theoretical composition; (C) percent abundances of individual taxa; (D) percent abundances of “unknown” taxa, i.e., not matching any
of the standard expected taxa.
For researchers using a custom mock community or mock communities from a different vendor, we
provide a step-by-step guide on preparing the training set as a FASTA file for full-length 16S rRNA gene
sequences of expected microbes using the DECIPHER R/BioC package. To this end, the taxonomic
assignment can be done using the assignTaxonomyCustomMock. We provide this tutorial on the package