Page 42 - Read Online
P. 42
Page 12 of 18 Fabbrini et al. Microbiome Res Rep 2023;2:25 https://dx.doi.org/10.20517/mrr.2023.25
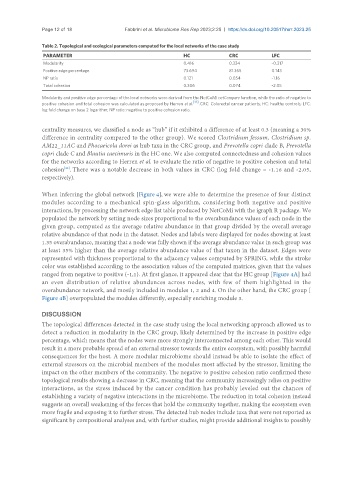
Table 2. Topological and ecological parameters computed for the local networks of the case study
PARAMETER HC CRC LFC
Modularity 0.416 0.334 -0.317
Positive edge percentage 73.694 81.365 0.143
NP ratio 0.121 0.054 -1.16
Total cohesion 0.306 0.074 -2.05
Modularity and positive edge percentage of the local networks were derived from the NetCoMi netCompare function, while the ratio of negative to
[33]
positive cohesion and total cohesion was calculated as proposed by Herren et al. . CRC: Colorectal cancer patients; HC: healthy controls; LFC:
log fold change on base 2 logarithm; NP ratio: negative to positive cohesion ratio.
centrality measures, we classified a node as “hub” if it exhibited a difference of at least 0.3 (meaning a 30%
difference in centrality compared to the other group). We scored Clostridium fessum, Clostridium sp.
AM22_11AC and Phocaeicola dorei as hub taxa in the CRC group, and Prevotella copri clade B, Prevotella
copri clade C and Blautia caecimuris in the HC one. We also computed connectedness and cohesion values
for the networks according to Herren et al. to evaluate the ratio of negative to positive cohesion and total
[33]
cohesion . There was a notable decrease in both values in CRC (log fold change = -1.16 and -2.05,
respectively).
When inferring the global network [Figure 4], we were able to determine the presence of four distinct
modules according to a mechanical spin-glass algorithm, considering both negative and positive
interactions, by processing the network edge list table produced by NetCoMi with the igraph R package. We
populated the network by setting node sizes proportional to the overabundance values of each node in the
given group, computed as the average relative abundance in that group divided by the overall average
relative abundance of that node in the dataset. Nodes and labels were displayed for nodes showing at least
1.35 overabundance, meaning that a node was fully shown if the average abundance value in such group was
at least 35% higher than the average relative abundance value of that taxon in the dataset. Edges were
represented with thickness proportional to the adjacency values computed by SPRING, while the stroke
color was established according to the association values of the computed matrices, given that the values
ranged from negative to positive (-1,1). At first glance, it appeared clear that the HC group [Figure 4A] had
an even distribution of relative abundances across nodes, with few of them highlighted in the
overabundance network, and mostly included in modules 1, 2 and 4. On the other hand, the CRC group [
Figure 4B] overpopulated the modules differently, especially enriching module 3.
DISCUSSION
The topological differences detected in the case study using the local networking approach allowed us to
detect a reduction in modularity in the CRC group, likely determined by the increase in positive edge
percentage, which means that the nodes were more strongly interconnected among each other. This would
result in a more probable spread of an external stressor towards the entire ecosystem, with possibly harmful
consequences for the host. A more modular microbiome should instead be able to isolate the effect of
external stressors on the microbial members of the modules most affected by the stressor, limiting the
impact on the other members of the community. The negative to positive cohesion ratio confirmed these
topological results showing a decrease in CRC, meaning that the community increasingly relies on positive
interactions, as the stress induced by the cancer condition has probably leveled out the chances of
establishing a variety of negative interactions in the microbiome. The reduction in total cohesion instead
suggests an overall weakening of the forces that hold the community together, making the ecosystem even
more fragile and exposing it to further stress. The detected hub nodes include taxa that were not reported as
significant by compositional analyses and, with further studies, might provide additional insights to possibly