Page 21 - Read Online
P. 21
Oquendo et al. J Transl Genet Genom 2021;5:89-111 https://dx.doi.org/10.20517/jtgg.2021.04 Page 93
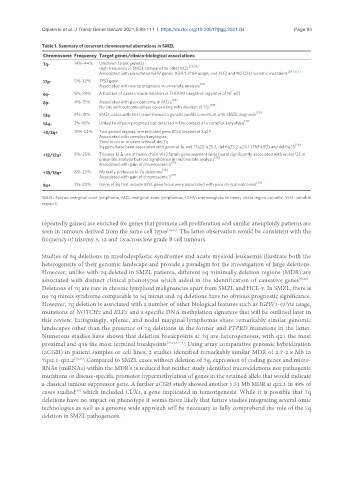
Table 1. Summary of recurrent chromosomal aberrations in SMZL
Chromosome Frequency Target genes/clinico-biological associations
7q- 14%-44% Unknown target gene(s)
High frequency in SMZL compared to other MZL [29,36]
[24,31,37]
Associated with unmutated IGHV genes, IGHV1-2*04 usage, and KLF2 and NOTCH2 somatic mutations
17p- 5%-32% TP53 gene
Associated with worse prognosis in univariate analysis [31]
6q- 8%-24% A fraction of cases include deletion of TNFAIP3 (negative regulator of NF-κB)
8p- 4%-15% Associated with poor outcome in MZLs [29]
No link with outcome unless co-existing with deletion of 17p [29]
13q- 5%-18% SMZL cases with this lesion showed a genetic profile consistent with SMZL diagnosis [29]
[31]
14q- 3%-10% Linked to inferior prognosis but detected in the context of a complex karyotype
+3/3q+ 15%-34% Two gained regions, one included gene BCL6 located at 3q27
Associated with complex karyotypes
Tend to occur in cases without del 7q
[29]
3q gains have been associated with gains at 1q and 17q22-q25.3, del 6q23.2-q24.1 (TNFAIP3) and del 6q25
+12/12q+ 8%-25% Trisomy 12 & use of heavy chain VH3 family gene segment were found significantly associated with worse OS in
univariate analysis but lost significance in multivariate analysis [36]
Associated with gain of chromosome 3 [31]
+18/18q+ 8%-23% Mutually exclusive to 7q deletions [36]
Associated with gain of chromosome 3 [31]
8q+ 2%-20% Gains of 8q that include MYC gene locus were associated with poor clinical outcomes [38]
SMZL: Splenic marginal zone lymphoma; MZL: marginal zone lymphomas; IGHV: immunoglobulin heavy chain region variable; VH3: variable
region 3.
repeatedly gained are enriched for genes that promote cell proliferation and similar aneuploidy patterns are
seen in tumours derived from the same cell types [34,35] . The latter observation would be consistent with the
frequency of trisomy 3, 12 and 18 across low grade B cell tumours.
Studies of 5q deletions in myelodysplastic syndromes and acute myeloid leukaemia illustrate both the
heterogeneity of their genomic landscape and provide a paradigm for the investigation of large deletions.
However, unlike with 7q deleted in SMZL patients, different 5q minimally deletion regions (MDR) are
associated with distinct clinical phenotypes which aided in the identification of causative genes [39,40] .
Deletions of 7q are rare in chronic lymphoid malignancies apart from SMZL and HCL-v. In SMZL, there is
no 7q minus syndrome comparable to 5q minus and 7q deletions have no obvious prognostic significance.
However, 7q deletion is associated with a number of other biological features such as IGHV1-02*04 usage,
mutations of NOTCH2 and KLF2 and a specific DNA methylation signature that will be outlined later in
this review. Intriguingly, splenic, and nodal marginal lymphomas share remarkably similar genomic
landscapes other than the presence of 7q deletions in the former and PTPRD mutations in the latter.
Numerous studies have shown that deletion breakpoints at 7q are heterogeneous, with q21 the most
proximal and q36 the most terminal breakpoints [19,31,41-45] . Using array comparative genomic hybridization
(aCGH) in patient samples or cell lines, 2 studies identified remarkably similar MDR of 2.7-2.8 Mb in
7q32.1-q32.2 [36,38] . Compared to SMZL cases without deletion of 7q, expression of coding genes and micro-
RNAs (miRNAs) within the MDR’s is reduced but neither study identified microdeletions nor pathogenic
mutations or disease-specific promotor hypermethylation of genes in the retained allele that would indicate
a classical tumour suppressor gene. A further aCGH study showed another 1.51 Mb MDR at q22.1 in 49% of
cases studied which included CUX1, a gene implicated in tumorigenesis. While it is possible that 7q
[46]
deletions have no impact on phenotype it seems more likely that future studies integrating several omic
technologies as well as a genome wide approach will be necessary to fully comprehend the role of the 7q
deletion in SMZL pathogenesis.