Page 117 - Read Online
P. 117
Page 330 Jiang et al. J Transl Genet Genom 2021;5:323-40 https://dx.doi.org/10.20517/jtgg.2021.21
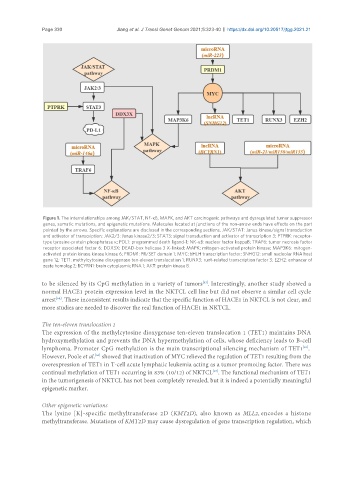
Figure 1. The interrelationships among JAK/STAT, NF-κB, MAPK, and AKT carcinogenic pathways and dysregulated tumor suppressor
genes, somatic mutations, and epigenetic mutations. Molecules located at junctions of the non-arrow ends have effects on the part
pointed by the arrows. Specific explanations are disclosed in the corresponding sections. JAK/STAT: Janus kinase/signal transduction
and activator of transcription; JAK2/3: Janus kinase2/3; STAT3: signal transduction and activator of transcription 3; PTPRK: receptor-
type tyrosine-protein phosphatase κ; PDL1: programmed death ligand-1; NK-κB: nuclear factor kappaB; TRAF6: tumor necrosis factor
receptor associated factor 6; DDX3X: DEAD-box helicase 3 X-linked; MAPK: mitogen-activated protein kinase; MAP3K6: mitogen-
activated protein kinase kinase kinase 6; PRDM1: PR/SET domain 1; MYC: bHLH transcription factor; SNHG12: small nucleolar RNA host
gene 12; TET1: methylcytosine dioxygenase ten-eleven translocation 1; RUNX3: runt-related transcription factor 3; EZH2: enhancer of
zeste homolog 2; BCYRN1: brain cytoplasmic RNA 1; AKT: protein kinase B.
to be silenced by its CpG methylation in a variety of tumors . Interestingly, another study showed a
[63]
normal HACE1 protein expression level in the NKTCL cell line but did not observe a similar cell cycle
arrest . These inconsistent results indicate that the specific function of HACE1 in NKTCL is not clear, and
[64]
more studies are needed to discover the real function of HACE1 in NKTCL.
The ten-eleven translocation 1
The expression of the methylcytosine dioxygenase ten-eleven translocation 1 (TET1) maintains DNA
hydroxymethylation and prevents the DNA hypermethylation of cells, whose deficiency leads to B-cell
[65]
lymphoma. Promoter CpG methylation is the main transcriptional silencing mechanism of TET1 .
However, Poole et al. showed that inactivation of MYC relieved the regulation of TET1 resulting from the
[66]
overexpression of TET1 in T-cell acute lymphatic leukemia acting as a tumor promoting factor. There was
continual methylation of TET1 occurring in 83% (10/12) of NKTCL . The functional mechanism of TET1
[67]
in the tumorigenesis of NKTCL has not been completely revealed, but it is indeed a potentially meaningful
epigenetic marker.
Other epigenetic variations
The lysine [K]-specific methyltransferase 2D (KMT2D), also known as MLL2, encodes a histone
methyltransferase. Mutations of KMT2D may cause dysregulation of gene transcription regulation, which