Page 40 - Read Online
P. 40
Page 4 of 10 Boshe et al. J Transl Genet Genom 2018;2:12. I https://doi.org/10.20517/jtgg.2018.18
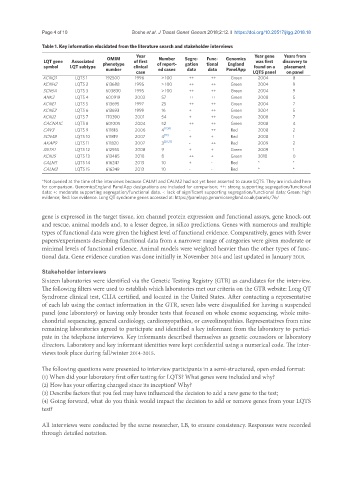
Table 1. Key information elucidated from the literature search and stakeholder interviews
Year Year gene Years from
LQT gene Associated OMIM of first Number Segre- Func- Genomics was first discovery to
symbol LQT subtype phenotype clinical of report- gation tional England found on a placement
number case ed cases data data PanelApp LQTS panel on panel
KCNQ1 LQTS 1 192500 1996 > 100 ++ ++ Green 2004 8
KCNH2 LQTS 2 613688 1995 > 100 ++ ++ Green 2004 9
SCN5A LQTS 3 603830 1995 > 100 ++ ++ Green 2004 9
ANK2 LQTS 4 600919 2003 57 ++ ++ Green 2008 5
KCNE1 LQTS 5 613695 1997 25 ++ ++ Green 2004 7
KCNE2 LQTS 6 613693 1999 16 + ++ Green 2004 5
KCNJ2 LQTS 7 170390 2001 54 + ++ Green 2008 7
CACNA1C LQTS 8 601005 2004 52 ++ ++ Green 2008 4
CAV3 LQTS 9 611818 2006 4 [17,18] - ++ Red 2008 2
SCN4B LQTS 10 611819 2007 4 [19] + + Red 2008 1
AKAP9 LQTS 11 611820 2007 3 [20,21] - ++ Red 2009 2
SNTA1 LQTS 12 612955 2008 9 + + Green 2009 1
KCNJ5 LQTS 13 613485 2010 8 ++ + Green 2010 0
CALM1 LQTS 14 616247 2013 10 + - Red * *
CALM2 LQTS 15 616249 2013 10 - - Red * *
*Not queried at the time of the interviews because CALM1 and CALM2 had not yet been asserted to cause LQTS. They are included here
for comparison. GenomicsEngland PanelApp designations are included for comparison; ++: strong supporting segregation/functional
data; +: moderate supporting segregation/functional data. -: lack of significant supporting segregation/functional data; Green: high
evidence; Red: low evidence. Long QT syndrome genes accessed at: https://panelapp.genomicsengland.co.uk/panels/76/
gene is expressed in the target tissue, ion channel protein expression and functional assays, gene knock-out
and rescue, animal models and, to a lesser degree, in silico predictions. Genes with numerous and multiple
types of functional data were given the highest level of functional evidence. Comparatively, genes with fewer
papers/experiments describing functional data from a narrower range of categories were given moderate or
minimal levels of functional evidence. Animal models were weighted heavier than the other types of func-
tional data. Gene evidence curation was done initially in November 2014 and last updated in January 2018.
Stakeholder interviews
Sixteen laboratories were identified via the Genetic Testing Registry (GTR) as candidates for the interview.
The following filters were used to establish which laboratories met our criteria on the GTR website: Long QT
Syndrome clinical test, CLIA certified, and located in the United States. After contacting a representative
of each lab using the contact information in the GTR, seven labs were disqualified for having a suspended
panel (one laboratory) or having only broader tests that focused on whole exome sequencing, whole mito-
chondrial sequencing, general cardiology, cardiomyopathies, or caveolinopathies. Representatives from nine
remaining laboratories agreed to participate and identified a key informant from the laboratory to partici-
pate in the telephone interviews. Key informants described themselves as genetic counselors or laboratory
directors. Laboratory and key informant identities were kept confidential using a numerical code. The inter-
views took place during fall/winter 2014-2015.
The following questions were presented to interview participants in a semi-structured, open ended format:
(1) When did your laboratory first offer testing for LQTS? What genes were included and why?
(2) How has your offering changed since its inception? Why?
(3) Describe factors that you feel may have influenced the decision to add a new gene to the test;
(4) Going forward, what do you think would impact the decision to add or remove genes from your LQTS
test?
All interviews were conducted by the same researcher, LB, to ensure consistency. Responses were recorded
through detailed notation.