Page 74 - Read Online
P. 74
Fraser. J Transl Genet Genom 2018;2:21. I https://doi.org/10.20517/jtgg.2018.27 Page 11 of 15
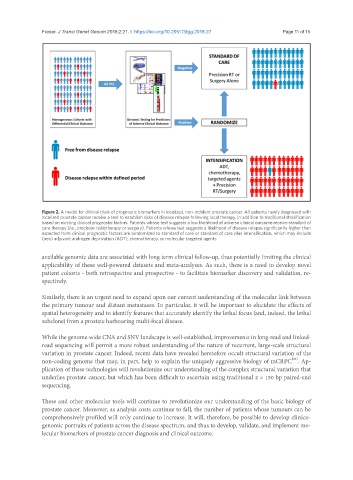
Figure 2. A model for clinical trials of prognostic biomarkers in localized, non-indolent prostate cancer. All patients newly diagnosed with
localized prostate cancer receive a test to establish risks of disease relapse following local therapy, in addition to traditional stratification
based on existing clinical prognostic factors. Patients whose test suggests a low likelihood of adverse clinical outcome receive standard of
care therapy (i.e., precision radiotherapy or surgery). Patients whose test suggests a likelihood of disease relapse significantly higher than
expected from clinical prognostic factors are randomized to standard of care or standard of care plus intensification, which may include
(neo) adjuvant androgen deprivation (ADT), chemotherapy, or molecular targeted agents
available genomic data are associated with long term clinical follow-up, thus potentially limiting the clinical
applicability of these well-powered datasets and meta-analyses. As such, there is a need to develop novel
patient cohorts - both retrospective and prospective - to facilitate biomarker discovery and validation, re-
spectively.
Similarly, there is an urgent need to expand upon our current understanding of the molecular link between
the primary tumour and distant metastases. In particular, it will be important to elucidate the effects of
spatial heterogeneity and to identify features that accurately identify the lethal focus (and, indeed, the lethal
subclone) from a prostate harbouring multi-focal disease.
While the genome-wide CNA and SNV landscape is well-established, improvements in long-read and linked-
read sequencing will permit a more robust understanding of the nature of recurrent, large-scale structural
variation in prostate cancer. Indeed, recent data have revealed heretofore occult structural variation of the
[86]
non-coding genome that may, in part, help to explain the uniquely aggressive biology of mCRPC . Ap-
plication of these technologies will revolutionize our understanding of the complex structural variation that
underlies prostate cancer, but which has been difficult to ascertain using traditional 2 × 150 bp paired-end
sequencing.
These and other molecular tools will continue to revolutionize our understanding of the basic biology of
prostate cancer. Moreover, as analysis costs continue to fall, the number of patients whose tumours can be
comprehensively profiled will only continue to increase. It will, therefore, be possible to develop clinico-
genomic portraits of patients across the disease spectrum, and thus to develop, validate, and implement mo-
lecular biomarkers of prostate cancer diagnosis and clinical outcome.