Page 52 - Read Online
P. 52
Page 46 Loong et al. J Transl Genet Genom 2023;7:27-49 https://dx.doi.org/10.20517/jtgg.2022.20
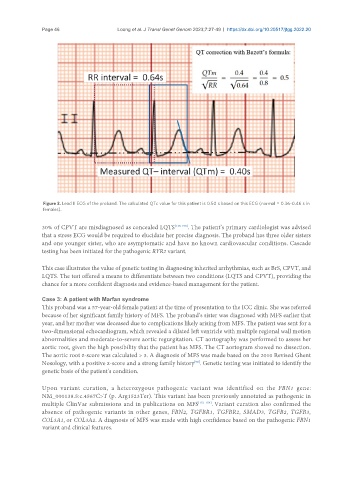
Figure 3. Lead II ECG of the proband. The calculated QTc value for this patient is 0.50 s based on this ECG (normal = 0.36-0.46 s in
females).
30% of CPVT are misdiagnosed as concealed LQTS [148-150] . The patient’s primary cardiologist was advised
that a stress ECG would be required to elucidate her precise diagnosis. The proband has three older sisters
and one younger sister, who are asymptomatic and have no known cardiovascular conditions. Cascade
testing has been initiated for the pathogenic RYR2 variant.
This case illustrates the value of genetic testing in diagnosing inherited arrhythmias, such as BrS, CPVT, and
LQTS. The test offered a means to differentiate between two conditions (LQTS and CPVT), providing the
chance for a more confident diagnosis and evidence-based management for the patient.
Case 3: A patient with Marfan syndrome
This proband was a 57-year-old female patient at the time of presentation to the ICC clinic. She was referred
because of her significant family history of MFS. The proband’s sister was diagnosed with MFS earlier that
year, and her mother was deceased due to complications likely arising from MFS. The patient was sent for a
two-dimensional echocardiogram, which revealed a dilated left ventricle with multiple regional wall motion
abnormalities and moderate-to-severe aortic regurgitation. CT aortography was performed to assess her
aortic root, given the high possibility that the patient has MFS. The CT aortogram showed no dissection.
The aortic root z-score was calculated > 3. A diagnosis of MFS was made based on the 2010 Revised Ghent
Nosology, with a positive z-score and a strong family history . Genetic testing was initiated to identify the
[98]
genetic basis of the patient’s condition.
Upon variant curation, a heterozygous pathogenic variant was identified on the FBN1 gene:
NM_000138.5:c.4567C>T (p. Arg1523Ter). This variant has been previously annotated as pathogenic in
multiple ClinVar submissions and in publications on MFS [151-154] . Variant curation also confirmed the
absence of pathogenic variants in other genes, FBN2, TGFBR1, TGFBR2, SMAD3, TGFB2, TGFB3,
COL3A1, or COL3A2. A diagnosis of MFS was made with high confidence based on the pathogenic FBN1
variant and clinical features.