Page 137 - Read Online
P. 137
Grelet et al. J Cancer Metastasis Treat 2019;5:16 I http://dx.doi.org/10.20517/2394-4722.2018.85 Page 7 of 10
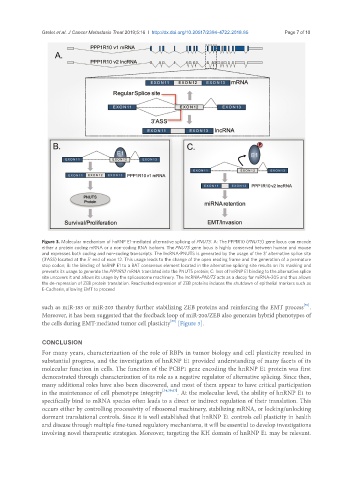
Figure 3. Molecular mechanism of hnRNP E1-mediated alternative splicing of PNUTS. A: The PPP1R10 (PNUTS) gene locus can encode
either a protein coding mRNA or a non-coding RNA isoform. The PNUTS gene locus is highly conserved between human and mouse
and expresses both coding and non-coding transcripts. The lncRNA-PNUTS is generated by the usage of the 3’ alternative splice site
(3’ASS) located at the 5’ end of exon 12. This usage leads to the change of the open reading frame and the generation of a premature
stop codon; B: the binding of hnRNP E1 to a BAT consensus element located in the alternative splicing site results on its masking and
prevents its usage to generate the PPP1R10 mRNA translated into the PNUTS protein; C: loss of hnRNP E1 binding to the alternative splice
site uncovers it and allows its usage by the spliceosome machinery. The lncRNA-PNUTS acts as a decoy for miRNA-205 and thus allows
the de-repression of ZEB protein translation. Reactivated expression of ZEB proteins induces the shutdown of epithelial markers such as
E-Cadherin, allowing EMT to proceed
[30]
such as miR-183 or miR-203 thereby further stabilizing ZEB proteins and reinforcing the EMT process .
Moreover, it has been suggested that the feedback loop of miR-200/ZEB also generates hybrid phenotypes of
[66]
the cells during EMT-mediated tumor cell plasticity [Figure 3].
CONCLUSION
For many years, characterization of the role of RBPs in tumor biology and cell plasticity resulted in
substantial progress, and the investigation of hnRNP E1 provided understanding of many facets of its
molecular function in cells. The function of the PCBP1 gene encoding the hnRNP E1 protein was first
demonstrated through characterization of its role as a negative regulator of alternative splicing. Since then,
many additional roles have also been discovered, and most of them appear to have critical participation
in the maintenance of cell phenotype integrity [34,39,67] . At the molecular level, the ability of hnRNP E1 to
specifically bind to mRNA species often leads to a direct or indirect regulation of their translation. This
occurs either by controlling processivity of ribosomal machinery, stabilizing mRNA, or locking/unlocking
dormant translational controls. Since it is well established that hnRNP E1 controls cell plasticity in health
and disease through multiple fine-tuned regulatory mechanisms, it will be essential to develop investigations
involving novel therapeutic strategies. Moreover, targeting the KH domain of hnRNP E1 may be relevant.