Page 73 - Read Online
P. 73
Page 6 of 19 Shi et al. J Cancer Metastasis Treat 2018;4:47 I http://dx.doi.org/10.20517/2394-4722.2018.32
A B
C
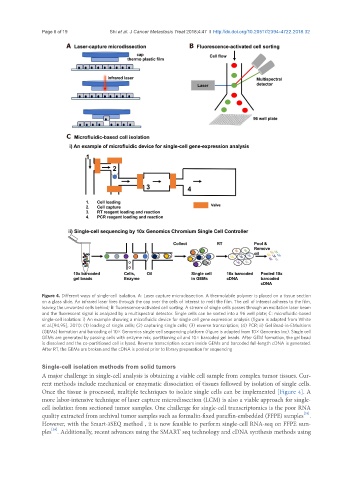
Figure 4. Different ways of single-cell isolation. A: Laser capture microdissection. A thermolabile polymer is placed on a tissue section
on a glass slide. An infrared laser fires through the cap over the cells of interest to melt the film. The cell of interest adheres to the film,
leaving the unwanted cells behind; B: fluorescence-activated cell sorting. A stream of single cells passes through an excitation laser beam
and the fluorescent signal is analyzed by a multispectral detector. Single cells can be sorted into a 96 well plate; C: microfluidic-based
single-cell isolation: i) An example showing a microfluidic device for single cell gene expression analysis (figure is adapted from White
et al.[94,95], 2011): (1) loading of single cells; (2) capturing single cells; (3) reverse transcription; (4) PCR; ii) Gel Bead-in-EMulsions
(GEMs) formation and barcoding of 10× Genomics single-cell sequencing platform (figure is adapted from 10× Genomics Inc). Single cell
GEMs are generated by passing cells with enzyme mix, partitioning oil and 10× barcoded gel beads. After GEM formation, the gel bead
is dissolved and the co-partitioned cell is lysed. Reverse transcription occurs inside GEMs and barcoded full-length cDNA is generated.
After RT, the GEMs are broken and the cDNA is pooled prior to library preparation for sequencing
Single-cell isolation methods from solid tumors
A major challenge in single-cell analysis is obtaining a viable cell sample from complex tumor tissues. Cur-
rent methods include mechanical or enzymatic dissociation of tissues followed by isolation of single cells.
Once the tissue is processed, multiple techniques to isolate single cells can be implemented [Figure 4]. A
more labor-intensive technique of laser capture microdissection (LCM) is also a viable approach for single-
cell isolation from sectioned tumor samples. One challenge for single-cell transcriptomics is the poor RNA
[39]
quality extracted from archival tumor samples such as formalin-fixed paraffin-embedded (FFPE) samples .
However, with the Smart-3SEQ method , it is now feasible to perform single-cell RNA-seq on FFPE sam-
[39]
ples . Additionally, recent advances using the SMART seq technology and cDNA synthesis methods using