Page 72 - Read Online
P. 72
Shi et al. J Cancer Metastasis Treat 2018;4:47 I http://dx.doi.org/10.20517/2394-4722.2018.32 Page 5 of 19
N
D
T
R
S
A
C
F
M
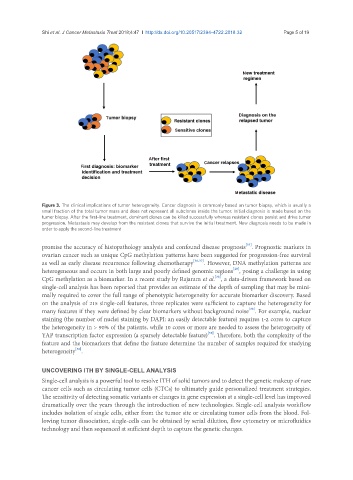
Figure 3. The clinical implications of tumor heterogeneity. Cancer diagnosis is commonly based on tumor biopsy, which is usually a
small fraction of the total tumor mass and does not represent all subclones inside the tumor. Initial diagnosis is made based on the
tumor biopsy. After the first-line treatment, dominant clones can be killed successfully whereas resistant clones persist and drive tumor
progression. Metastasis may develop from the resistant clones that survive the initial treatment. New diagnosis needs to be made in
order to apply the second-line treatment
[35]
promise the accuracy of histopathology analysis and confound disease prognosis . Prognostic markers in
ovarian cancer such as unique CpG methylation patterns have been suggested for progression-free survival
as well as early disease recurrence following chemotherapy [36,37] . However, DNA methylation patterns are
[20]
heterogeneous and occurs in both large and poorly defined genomic regions , posing a challenge in using
[38]
CpG methylation as a biomarker. In a recent study by Rajaram et al. , a data-driven framework based on
single-cell analysis has been reported that provides an estimate of the depth of sampling that may be mini-
mally required to cover the full range of phenotypic heterogeneity for accurate biomarker discovery. Based
on the analysis of 215 single-cell features, three replicates were sufficient to capture the heterogeneity for
[38]
many features if they were defined by clear biomarkers without background noise . For example, nuclear
staining (the number of nuclei staining by DAPI: an easily detectable feature) requires 1-2 cores to capture
the heterogeneity in > 90% of the patients, while 10 cores or more are needed to assess the heterogeneity of
[38]
YAP transcription factor expression (a sparsely detectable feature) . Therefore, both the complexity of the
feature and the biomarkers that define the feature determine the number of samples required for studying
[38]
heterogeneity .
UNCOVERING ITH BY SINGLE-CELL ANALYSIS
Single-cell analysis is a powerful tool to resolve ITH of solid tumors and to detect the genetic makeup of rare
cancer cells such as circulating tumor cells (CTCs) to ultimately guide personalized treatment strategies.
The sensitivity of detecting somatic variants or changes in gene expression at a single-cell level has improved
dramatically over the years through the introduction of new technologies. Single-cell analysis workflow
includes isolation of single cells, either from the tumor site or circulating tumor cells from the blood. Fol-
lowing tumor dissociation, single-cells can be obtained by serial dilution, flow cytometry or microfluidics
technology and then sequenced at sufficient depth to capture the genetic changes.