Page 936 - Read Online
P. 936
Caron de Fromentel et al. Hepatoma Res 2020;6:80 I http://dx.doi.org/10.20517/2394-5079.2020.77 Page 5 of 18
A
B
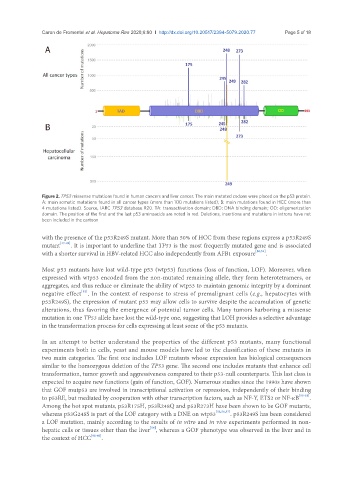
Figure 2. TP53 missense mutations found in human cancers and liver cancer. The main mutated codons were placed on the p53 protein.
A: main somatic mutations found in all cancer types (more than 100 mutations listed); B: main mutations found in HCC (more than
4 mutations listed). Source, IARC TP53 database R20. TA: transactivation domain; DBD: DNA binding domain; OD: oligomerization
domain. The position of the first and the last p53 aminoacids are noted in red. Deletions, insertions and mutations in introns have not
been included in the cartoon
with the presence of the p53R249S mutant. More than 50% of HCC from these regions express a p53R249S
mutant [47-49] . It is important to underline that TP53 is the most frequently mutated gene and is associated
with a shorter survival in HBV-related HCC also independently from AFB1 exposure [50,51] .
Most p53 mutants have lost wild-type p53 (wtp53) functions (loss of function, LOF). Moreover, when
expressed with wtp53 encoded from the non-mutated remaining allele, they form heterotetramers, or
aggregates, and thus reduce or eliminate the ability of wtp53 to maintain genomic integrity by a dominant
[52]
negative effect . In the context of response to stress of premalignant cells (e.g., hepatocytes with
p53R249S), the expression of mutant p53 may allow cells to survive despite the accumulation of genetic
alterations, thus favoring the emergence of potential tumor cells. Many tumors harboring a missense
mutation in one TP53 allele have lost the wild-type one, suggesting that LOH provides a selective advantage
in the transformation process for cells expressing at least some of the p53 mutants.
In an attempt to better understand the properties of the different p53 mutants, many functional
experiments both in cells, yeast and mouse models have led to the classification of these mutants in
two main categories. The first one includes LOF mutants whose expression has biological consequences
similar to the homozygous deletion of the TP53 gene. The second one includes mutants that enhance cell
transformation, tumor growth and aggressiveness compared to their p53-null counterparts. This last class is
expected to acquire new functions (gain of function, GOF). Numerous studies since the 1990s have shown
that GOF mutp53 are involved in transcriptional activation or repression, independently of their binding
to p53RE, but mediated by cooperation with other transcription factors, such as NF-Y, ETS2 or NF-kB [53-55] .
Among the hot spot mutants, p53R175H, p53R248Q and p53R273H have been shown to be GOF mutants,
whereas p53G245S is part of the LOF category with a DNE on wtp53 [53,56,57] . p53R249S has been considered
a LOF mutation, mainly according to the results of in vitro and in vivo experiments performed in non-
[56]
hepatic cells or tissues other than the liver , whereas a GOF phenotype was observed in the liver and in
the context of HCC [58-60] .