Page 17 - Read Online
P. 17
Page 6 of 12 Nishioka et al. Hepatoma Res 2018;4:1 I http://dx.doi.org/10.20517/2394-5079.2017.46
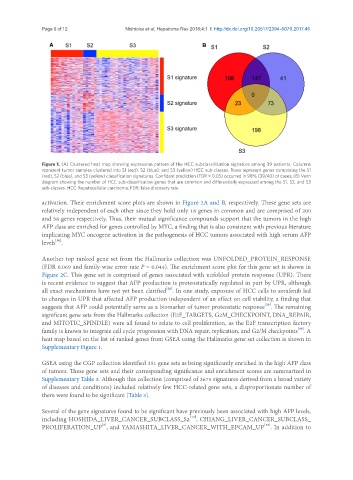
A B
Figure 1. (A) Clustered heat map showing expression pattern of the HCC subclassification signature among 39 patients. Columns
represent tumor samples clustered into S1 (red), S2 (blue), and S3 (yellow) HCC sub-classes. Rows represent genes comprising the S1
(red), S2 (blue), and S3 (yellow) classification signatures. Confident prediction (FDR < 0.05) occurred in 98% (39/40) of cases; (B) Venn
diagram showing the number of HCC sub-classification genes that are common and differentially expressed among the S1, S2, and S3
sub-classes. HCC: hepatocellular carcinoma; FDR: false discovery rate
activation. Their enrichment score plots are shown in Figure 2A and B, respectively. These gene sets are
relatively independent of each other since they hold only 18 genes in common and are comprised of 200
and 58 genes respectively. Thus, their mutual significance compounds support that the tumors in the high
AFP class are enriched for genes controlled by MYC, a finding that is also consistent with previous literature
implicating MYC oncogene activation in the pathogenesis of HCC tumors associated with high serum AFP
[10]
levels .
Another top ranked gene set from the Hallmarks collection was UNFOLDED_PROTEIN_RESPONSE
(FDR 0.069 and family-wise error rate P = 0.044). The enrichment score plot for this gene set is shown in
Figure 2C. This gene set is comprised of genes associated with unfolded protein response (UPR). There
is recent evidence to suggest that AFP production is proteostatically regulated in part by UPR, although
[28]
all exact mechanisms have not yet been clarified . In one study, exposure of HCC cells to sorafenib led
to changes in UPR that affected AFP production independent of an effect on cell viability, a finding that
[29]
suggests that AFP could potentially serve as a biomarker of tumor proteostatic response . The remaining
significant gene sets from the Hallmarks collection (E2F_TARGETS, G2M_CHECKPOINT, DNA_REPAIR,
and MITOTIC_SPINDLE) were all found to relate to cell proliferation, as the E2F transcription factory
family is known to integrate cell cycle progression with DNA repair, replication, and G2/M checkpoints . A
[30]
heat map based on the list of ranked genes from GSEA using the Hallmarks gene set collection is shown in
Supplementary Figure 1.
GSEA using the CGP collection identified 351 gene sets as being significantly enriched in the high AFP class
of tumors. These gene sets and their corresponding significance and enrichment scores are summarized in
Supplementary Table 3. Although this collection (comprised of 2675 signatures derived from a broad variety
of diseases and conditions) included relatively few HCC-related gene sets, a disproportionate number of
them were found to be significant [Table 3].
Several of the gene signatures found to be significant have previously been associated with high AFP levels,
[10]
including HOSHIDA_LIVER_CANCER_SUBCLASS_S2 , CHIANG_LIVER_CANCER_SUBCLASS_
[12]
PROLIFERATION_UP , and YAMASHITA_LIVER_CANCER_WITH_EPCAM_UP . In addition to
[8]