Page 105 - Read Online
P. 105
Link et al. Roles of p53 in extrinsic factor-induced liver carcinogenesis
A
B
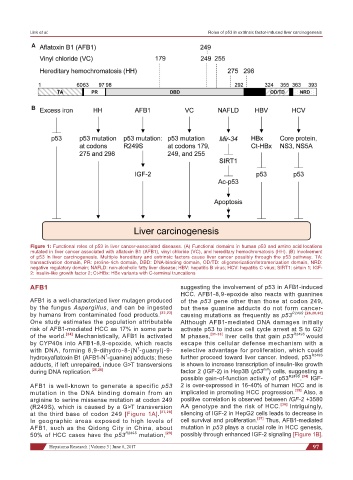
Figure 1: Functional roles of p53 in liver cancer-associated diseases. (A) Functional domains in human p53 and amino acid locations
mutated in liver cancer associated with aflatoxin B1 (AFB1), vinyl chloride (VC), and hereditary hemochromatosis (HH). (B) Involvement
of p53 in liver carcinogenesis. Multiple hereditary and extrinsic factors cause liver cancer possibly through the p53 pathway. TA:
transactivation domain, PR: proline-rich domain, DBD: DNA-binding domain, OD/TD: oligomerization/tetramerization domain, NRD:
negative regulatory domain; NAFLD: non-alcoholic fatty liver disease; HBV: hepatitis B virus; HCV: hepatitis C virus; SIRT1: sirtuin 1; IGF-
2: insulin-like growth factor 2; Ct-HBx: HBx variants with C-terminal truncations
AFB1 suggesting the involvement of p53 in AFB1-induced
HCC. AFB1-8,9-epoxide also reacts with guanines
AFB1 is a well-characterized liver mutagen produced of the p53 gene other than those at codon 249,
by the fungus Aspergillus, and can be ingested but these guanine adducts do not form cancer-
.
by humans from contaminated food products. [22,23] causing mutations as frequently as p53 R249S [26,28,30]
One study estimates the population attributable Although AFB1-mediated DNA damages initially
risk of AFB1-mediated HCC as 17% in some parts activate p53 to induce cell cycle arrest at S to G2/
of the world. [24] Mechanistically, AFB1 is activated M phases, [31-33] liver cells that gain p53 R249S would
by CYP40s into AFB1-8,9-epoxide, which reacts escape this cellular defense mechanism with a
7
with DNA, forming 8,9-dihydro-8-(N -guanyl)-9- selective advantage for proliferation, which could
7
hydroxyaflatoxin B1 (AFB1-N -guanine) adducts; these further proceed toward liver cancer. Indeed, p53 R249S
adducts, if left unrepaired, induce G>T transversions is shown to increase transcription of insulin-like growth
null
during DNA replication. [25,26] factor 2 (IGF-2) in Hep3B (p53 ) cells, suggesting a
possible gain-of-function activity of p53 R249S [34] IGF-
.
AFB1 is well-known to generate a specific p53 2 is over-expressed in 16-40% of human HCC and is
mutation in the DNA binding domain from an implicated in promoting HCC progression. [35] Also, a
arginine to serine missense mutation at codon 249 positive correlation is observed between IGF-2 +3580
(R249S), which is caused by a G>T transversion AA genotype and the risk of HCC. [36] Intriguingly,
at the third base of codon 249 [Figure 1A]. [27,28] silencing of IGF-2 in HepG2 cells leads to decrease in
In geographic areas exposed to high levels of cell survival and proliferation. [37] Thus, AFB1-mediated
AFB1, such as the Qidong City in China, about mutation in p53 plays a crucial role in HCC genesis,
50% of HCC cases have the p53 R249S mutation, [29] possibly through enhanced IGF-2 signaling [Figure 1B].
Hepatoma Research ¦ Volume 3 ¦ June 6, 2017 97