Page 53 - Read Online
P. 53
Page 12 Biersack. Cancer Drug Resist 2019;2:1-17 I http://dx.doi.org/10.20517/cdr.2019.09
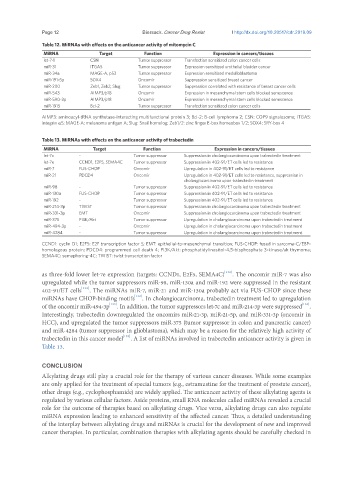
Table 12. MiRNAs with effects on the anticancer activity of mitomycin C
MiRNA Target Function Expression in cancers/tissues
let-7-1 CSN Tumor suppressor Transfection sensitized colon cancer cells
miR-31 ITGA5 Tumor suppressor Expression sensitized urothelial bladder cancer
miR-34a MAGE-A, p53 Tumor suppressor Expression sensitized medulloblastoma
miR-191-5p SOX4 Oncomir Suppression sensitized breast cancer
miR-200 Zeb1, Zeb2, Slug Tumor suppressor Suppression correlated with resistance of breast cancer cells
miR-543 AIMP3/p18 Oncomir Expression in mesenchymal stem cells blocked senescence
miR-590-3p AIMP3/p18 Oncomir Expression in mesenchymal stem cells blocked senescence
miR-1915 Bcl-2 Tumor suppressor Transfection sensitized colon cancer cells
AIMP3: aminoacyl-tRNA synthetase-interacting multifunctional protein 3; Bcl-2: B-cell lymphoma 2; CSN: COP9 signalosome; ITGA5:
integrin α5; MAGE-A: melanoma antigen A; Slug: Snail homolog; Zeb1/2: zinc finger E-box homeobox 1/2; SOX4: SRY-box 4
Table 13. MiRNAs with effects on the anticancer activity of trabectedin
MiRNA Target Function Expression in cancers/tissues
let-7c - Tumor suppressor Suppression in cholangiocarcinoma upon trabectedin treatment
let-7e CCND1, E2F5, SEMA4C Tumor suppressor Suppression in 402-91/ET cells led to resistance
miR-7 FUS-CHOP Oncomir Upregulation in 402-91/ET cells led to resistance
miR-21 PDCD4 Oncomir Upregulation in 402-91/ET cells led to resistance, suppression in
cholangiocarcinoma upon trabectedin treatment
miR-98 - Tumor suppressor Suppression in 402-91/ET cells led to resistance
miR-130a FUS-CHOP Tumor suppressor Suppression in 402-91/ET cells led to resistance
miR-192 - Tumor suppressor Suppression in 402-91/ET cells led to resistance
miR-214-3p TWIST Tumor suppressor Suppression in cholangiocarcinoma upon trabectedin treatment
miR-331-3p EMT Oncomir Suppression in cholangiocarcinoma upon trabectedin treatment
miR-375 PI3K/Akt Tumor suppressor Upregulation in cholangiocarcinoma upon trabectedin treatment
miR-494-3p - Oncomir Upregulation in cholangiocarcinoma upon trabectedin treatment
miR-4284 - Tumor suppressor Upregulation in cholangiocarcinoma upon trabectedin treatment
CCND1: cyclin D1; E2F5: E2F transcription factor 5; EMT: epithelial-to-mesenchymal transition; FUS-CHOP: fused in sarcoma-C/EBP-
homologous protein; PDCD4: programmed cell death 4; PI3K/Akt: phosphatidylinositol-4,5-bisphosphate 3-kinase/ak thymoma;
SEMA4C: semaphoring-4C; TWIST: twist transcription factor
as three-fold lower let-7e expression (targets: CCND1, E2F5, SEMA4C) [132] . The oncomir miR-7 was also
upregulated while the tumor suppressors miR-98, miR-130a and miR-192 were suppressed in the resistant
402-91/ET cells [132] . The miRNAs miR-7, miR-21 and miR-130a probably act via FUS-CHOP since these
[132]
miRNAs have CHOP-binding motifs . In cholangiocarcinoma, trabectedin treatment led to upregulation
[133]
[133]
of the oncomir miR-494-3p . In addition, the tumor suppressors let-7c and miR-214-3p were suppressed .
Interestingly, trabectedin downregulated the oncomirs miR-21-3p, miR-21-5p, and miR-331-3p (oncomir in
HCC), and upregulated the tumor suppressors miR-375 (tumor suppressor in colon and pancreatic cancer)
and miR-4284 (tumor suppressor in glioblastoma), which may be a reason for the relatively high activity of
trabectedin in this cancer model [133] . A list of miRNAs involved in trabectedin anticancer activity is given in
Table 13.
CONCLUSION
Alkylating drugs still play a crucial role for the therapy of various cancer diseases. While some examples
are only applied for the treatment of special tumors (e.g., estramustine for the treatment of prostate cancer),
other drugs (e.g., cyclophosphamide) are widely applied. The anticancer activity of these alkylating agents is
regulated by various cellular factors. Aside proteins, small RNA molecules called miRNAs revealed a crucial
role for the outcome of therapies based on alkylating drugs. Vice versa, alkylating drugs can also regulate
miRNA expression leading to enhanced sensitivity of the affected cancer. Thus, a detailed understanding
of the interplay between alkylating drugs and miRNAs is crucial for the development of new and improved
cancer therapies. In particular, combination therapies with alkylating agents should be carefully checked in