Page 79 - Read Online
P. 79
De Mattia et al. Cancer Drug Resist 2019;2:116-30 I http://dx.doi.org/10.20517/cdr.2019.04 Page 121
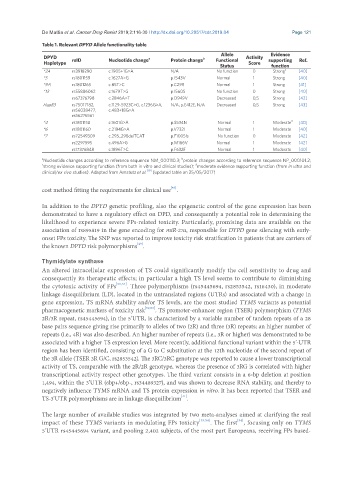
Table 1. Relevant DPYD Allele functionality table
Evidence
Allele
DPYD rsID Nucleotide change a Protein change b Functional Activity supporting Ref.
Haplotype Score
Status function
*2A rs3918290 c.1905+1G>A N/A No function 0 Strong c [40]
*5 rs1801159 c.1627A>G p.I543V Normal 1 Strong [40]
*9A rs1801265 c.85T>C p.C29R Normal 1 Strong [41]
*13 rs55886062 c.1679T>G p.I560S No function 0 Strong [40]
rs67376798 c.2846A>T p.D949V Decreased 0,5 Strong [42]
HapB3 rs75017182, c.1129-5923C>G, c.1236G>A, N/A, p.E412E, N/A Decreased 0,5 Strong [43]
rs56038477, c.483+18G>A
rs56276561
*4 rs1801158 c.1601G>A p.S534N Normal 1 Moderate d [40]
*6 rs1801160 c.2194G>A p.V732I Normal 1 Moderate [40]
*7 rs72549309 c.295_298delTCAT p.F100Sfs No function 0 Moderate [42]
rs2297595 c.496A>G p.M166V Normal 1 Moderate [42]
rs17376848 c.1896T>C p.F632F Normal 1 Moderate [40]
b
a Nucleotide changes according to reference sequence NM_000110.3; protein changes according to reference sequence NP_000101.2;
d
c strong evidence supporting function (from both in vitro and clinical studies); moderate evidence supporting function (from in vitro and
clinical/ex vivo studies). Adapted from Amstutz et al. [19] (updated table on 25/05/2017)
[48]
cost method fitting the requirements for clinical use .
In addition to the DPYD genetic profiling, also the epigenetic control of the gene expression has been
demonstrated to have a regulatory effect on DPD, and consequently a potential role in determining the
likelihood to experience severe FPs-related toxicity. Particularly, promising data are available on the
association of rs895819 in the gene encoding for miR-27a, responsible for DYPD gene silencing with early-
onset FPs toxicity. The SNP was reported to improve toxicity risk stratification in patients that are carriers of
[49]
the known DPYD risk polymorphisms .
Thymidylate synthase
An altered intracellular expression of TS could significantly modify the cell sensitivity to drug and
consequently its therapeutic effects; in particular a high TS level seems to contribute to diminishing
the cytotoxic activity of FPs [50,51] . Three polymorphisms (rs45445694, rs2853542, rs16430), in moderate
linkage disequilibrium (LD), located in the untranslated regions (UTRs) and associated with a change in
gene expression, TS mRNA stability and/or TS levels, are the most studied TYMS variants as potential
pharmacogenetic markers of toxicity risk [52,53] . TS promoter-enhancer region (TSER) polymorphism (TYMS
2R/3R repeat, rs45445694), in the 5’UTR, is characterized by a variable number of tandem repeats of a 28
base pairs sequence giving rise primarily to alleles of two (2R) and three (3R) repeats; an higher number of
repeats (i.e., 4R) was also described. An higher number of repeats (i.e., 3R or higher) was demonstrated to be
associated with a higher TS expression level. More recently, additional functional variant within the 5’-UTR
region has been identified, consisting of a G to C substitution at the 12th nucleotide of the second repeat of
the 3R allele (TSER 3R G/C, rs2853542). The 3RC/3RC genotype was reported to cause a lower transcriptional
activity of TS, comparable with the 2R/2R genotype, whereas the presence of 3RG is correlated with higher
transcriptional activity respect other genotypes. The third variant consists in a 6-bp deletion at position
1,494, within the 3’UTR (6bp+/6bp-, rs34489327), and was shown to decrease RNA stability, and thereby to
negatively influence TYMS mRNA and TS protein expression in vitro. It has been reported that TSER and
[41]
TS-3’UTR polymorphisms are in linkage disequilibrium .
The large number of available studies was integrated by two meta-analyses aimed at clarifying the real
[54]
impact of these TYMS variants in modulating FPs toxicity [39,54] . The first , focusing only on TYMS
5’UTR rs45445694 variant, and pooling 2,402 subjects, of the most part Europeans, receiving FPs based-