Page 52 - Read Online
P. 52
Page 230 Crisafulli et al. Cancer Drug Resist 2019;2:225-41 I http://dx.doi.org/10.20517/cdr.2018.008
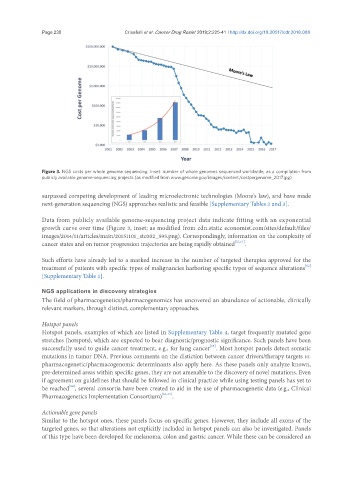
Figure 3. NGS costs per whole genome sequencing. Inset: number of whole genomes sequenced worldwide, as a compilation from
publicly available genome-sequencing projects (as modified from www.genome.gov/images/content/costpergenome_2017.jpg)
surpassed competing development of leading microelectronic technologies (Moore’s law), and have made
next-generation sequencing (NGS) approaches realistic and feasible [Supplementary Tables 2 and 3].
Data from publicly available genome-sequencing project data indicate fitting with an exponential
growth curve over time (Figure 3, inset; as modified from cdn.static economist.com/sites/default/files/
images/2014/11/articles/main/20151101_stc002_595.png). Correspondingly, information on the complexity of
cancer states and on tumor progression trajectories are being rapidly obtained [21,61] .
Such efforts have already led to a marked increase in the number of targeted therapies approved for the
[62]
treatment of patients with specific types of malignancies harboring specific types of sequence alterations
[Supplementary Table 1].
NGS applications in discovery strategies
The field of pharmacogenetics/pharmacogenomics has uncovered an abundance of actionable, clinically
relevant markers, through distinct, complementary approaches.
Hotspot panels
Hotspot panels, examples of which are listed in Supplementary Table 4, target frequently mutated gene
stretches (hotspots), which are expected to bear diagnostic/prognostic significance. Such panels have been
[63]
successfully used to guide cancer treatment, e.g., for lung cancer . Most hotspot panels detect somatic
mutations in tumor DNA. Previous comments on the distiction between cancer drivers/therapy targets vs.
pharmacogenetic/pharmacogenomic determinants also apply here. As these panels only analyze known,
pre-determined areas within specific genes, they are not amenable to the discovery of novel mutations. Even
if agreement on guidelines that should be followed in clinical practice while using testing panels has yet to
[64]
be reached , several consortia have been created to aid in the use of pharmacogenetic data (e.g., Clinical
Pharmacogenetics Implementation Consortium) [65-67] .
Actionable gene panels
Similar to the hotspot ones, these panels focus on specific genes. However, they include all exons of the
targeted genes, so that alterations not explicitly included in hotspot panels can also be investigated. Panels
of this type have been developed for melanoma, colon and gastric cancer. While these can be considered an