Page 51 - Read Online
P. 51
Crisafulli et al. Cancer Drug Resist 2019;2:225-41 I http://dx.doi.org/10.20517/cdr.2018.008 Page 229
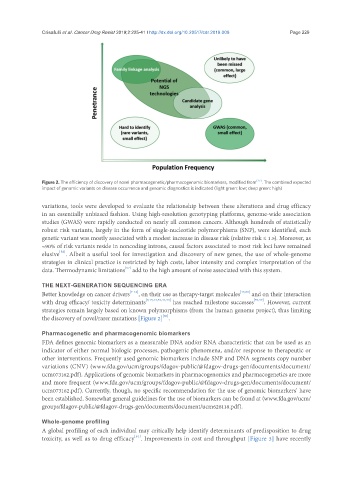
Figure 2. The efficiency of discovery of novel pharmacogenetic/pharmacogenomic biomarkers, modified from [27] . The combined expected
impact of genomic variants on disease occurrence and genomic diagnostics is indicated (light green: low; deep green: high)
variations, tools were developed to evaluate the relationship between these alterations and drug efficacy
in an essentially unbiased fashion. Using high-resolution genotyping platforms, genome-wide association
studies (GWAS) were rapidly conducted on nearly all common cancers. Although hundreds of statistically
robust risk variants, largely in the form of single-nucleotide polymorphisms (SNP), were identified, each
genetic variant was mostly associated with a modest increase in disease risk (relative risk ≤ 1.5). Moreover, as
≈90% of risk variants reside in noncoding introns, causal factors associated to most risk loci have remained
[56]
elusive . Albeit a useful tool for investigation and discovery of new genes, the use of whole-genome
strategies in clinical practice is restricted by high costs, labor intensity and complex interpretation of the
[57]
data. Thermodynamic limitations add to the high amount of noise associated with this system.
THE NEXT-GENERATION SEQUENCING ERA
Better knowledge on cancer drivers [7-14] , on their use as therapy-target molecules [13,58] and on their interaction
with drug efficacy/ toxicity determinants [9,13,14,39,41,59] has reached milestone successes [39,59] . However, current
strategies remain largely based on known polymorphisms (from the human genome project), thus limiting
[60]
the discovery of novel/rarer mutations [Figure 2] .
Pharmacogenetic and pharmacogenomic biomarkers
FDA defines genomic biomarkers as a measurable DNA and/or RNA characteristic that can be used as an
indicator of either normal biologic processes, pathogenic phenomena, and/or response to therapeutic or
other interventions. Frequently used genomic biomarkers include SNP and DNA segments copy number
variations (CNV) (www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/
ucm073162.pdf). Applications of genomic biomarkers in pharmacogenomics and pharmacogenetics are more
and more frequent (www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/
ucm073162.pdf). Currently, though, no specific recommendation for the use of genomic biomarkers’ have
been established. Somewhat general guidelines for the use of biomarkers can be found at (www.fda.gov/ucm/
groups/fdagov-public/@fdagov-drugs-gen/documents/document/ucm628118.pdf).
Whole-genome profiling
A global profiling of each individual may critically help identify determinants of predisposition to drug
[61]
toxicity, as well as to drug efficacy . Improvements in cost and throughput [Figure 3] have recently