Page 125 - Read Online
P. 125
Page 76 Shek et al. Cancer Drug Resist 2019;2:69-81 I http://dx.doi.org/10.20517/cdr.2018.20
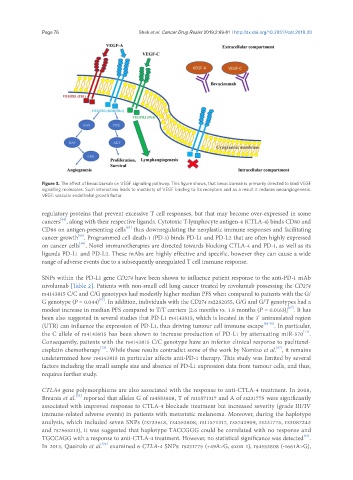
Figure 3. The effect of bevacizumab on VEGF signalling pathway. This figure shows, that bevacizumab is primarily directed to bind VEGF
signalling molecules. Such interaction leads to inactivity of VEGF binding to its receptors and as a result it reduces neoangiogenesis.
VEGF: vascular endothelial growth factor
regulatory proteins that prevent excessive T cell responses, but that may become over-expressed in some
[64]
cancers , along with their respective ligands. Cytotoxic T-lymphocyte antigen-4 (CTLA-4) binds CD80 and
[65]
CD86 on antigen-presenting cells thus downregulating the neoplastic immune responses and facilitating
[66]
cancer growth . Programmed cell death-1 (PD-1) binds PD-L1 and PD-L2 that are often highly expressed
[66]
on cancer cells . Novel immunotherapies are directed towards blocking CTLA-4 and PD-1, as well as its
ligands PD-L1 and PD-L2. These mAbs are highly effective and specific, however they can cause a wide
range of adverse events due to a subsequently unregulated T cell immune response.
SNPs within the PD-L1 gene CD274 have been shown to influence patient response to the anti-PD-1 mAb
nivolumab [Table 2]. Patients with non-small cell lung cancer treated by nivolumab possessing the CD274
rs4143815 C/C and C/G genotypes had modestly higher median PFS when compared to patients with the G/
[67]
G genotype (P = 0.044) . In addition, individuals with the CD274 rs2282055, G/G and G/T genotypes had a
[67]
modest increase in median PFS compared to T/T carriers [2.6 months vs. 1.8 months (P = 0.0163)] . It has
been also suggested in several studies that PD-L1 rs4143815, which is located in the 3’ untranslated region
(UTR) can influence the expression of PD-L1, thus driving tumour cell immune escape [68-70] . In particular,
the C allele of rs4143815 has been shown to increase production of PD-L1 by attenuating miR-570 .
[71]
Consequently, patients with the rs4143815 C/C genotype have an inferior clinical response to paclitaxel-
cisplatin chemotherapy . While these results contradict some of the work by Nomizo et al. , it remains
[67]
[72]
undetermined how rs4143815 in particular affects anti-PD-1 therapy. This study was limited by several
factors including the small sample size and absence of PD-L1 expression data from tumour cells, and thus,
requires further study.
CTLA4 gene polymorphisms are also associated with the response to anti-CTLA-4 treatment. In 2008,
Breunis et al. reported that alleles G of rs4553808, T of rs11571317 and A of rs231775 were significantly
[73]
associated with improved response to CTLA-4 blockade treatment but increased severity (grade III/IV
immune-related adverse events) in patients with metastatic melanoma. Moreover, during the haplotype
analysis, which included seven SNPs (rs733618, rs4553808, rs11571317, rs5742909, rs231775, rs3087243
and rs7565213), it was suggested that haplotype TACCGGG could be correlated with no response and
[73]
TGCCAGG with a response to anti-CTLA-4 treatment. However, no statistical significance was detected .
[74]
In 2013, Queirolo et al. examined 6 CTLA-4 SNPs: rs231775 (+49A>G, exon 1), rs4553808 (-1661A>G),