Page 76 - Read Online
P. 76
Aguiar. Rare Dis Orphan Drugs J 2024;3:13 https://dx.doi.org/10.20517/rdodj.2023.56 Page 17 of 29
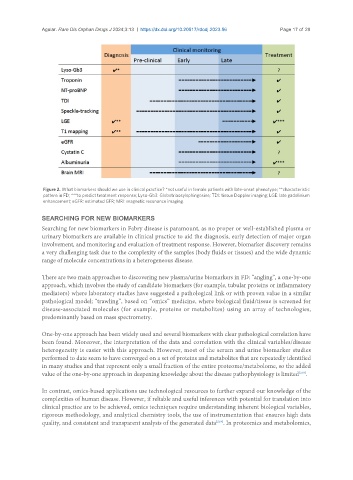
Figure 2. What biomarkers should we use in clinical practice? *not useful in female patients with late-onset phenotype; **characteristic
pattern in FD; ***to predict treatment response; Lyso-Gb3: Globotriaosylsphingosine; TDI: tissue Doppler imaging; LGE: late gadolinium
enhancement; eGFR: estimated GFR; MRI: magnetic resonance imaging.
SEARCHING FOR NEW BIOMARKERS
Searching for new biomarkers in Fabry disease is paramount, as no proper or well-established plasma or
urinary biomarkers are available in clinical practice to aid the diagnosis, early detection of major organ
involvement, and monitoring and evaluation of treatment response. However, biomarker discovery remains
a very challenging task due to the complexity of the samples (body fluids or tissues) and the wide dynamic
range of molecule concentrations in a heterogeneous disease.
There are two main approaches to discovering new plasma/urine biomarkers in FD: “angling”, a one-by-one
approach, which involves the study of candidate biomarkers (for example, tubular proteins or inflammatory
mediators) where laboratory studies have suggested a pathological link or with proven value in a similar
pathological model; “trawling”, based on “omics” medicine, where biological fluid/tissue is screened for
disease-associated molecules (for example, proteins or metabolites) using an array of technologies,
predominantly based on mass spectrometry.
One-by-one approach has been widely used and several biomarkers with clear pathological correlation have
been found. Moreover, the interpretation of the data and correlation with the clinical variables/disease
heterogeneity is easier with this approach. However, most of the serum and urine biomarker studies
performed to date seem to have converged on a set of proteins and metabolites that are repeatedly identified
in many studies and that represent only a small fraction of the entire proteome/metabolome, so the added
[218]
value of the one-by-one approach in deepening knowledge about the disease pathophysiology is limited .
In contrast, omics-based applications use technological resources to further expand our knowledge of the
complexities of human disease. However, if reliable and useful inferences with potential for translation into
clinical practice are to be achieved, omics techniques require understanding inherent biological variables,
rigorous methodology, and analytical chemistry tools, the use of instrumentation that ensures high data
quality, and consistent and transparent analysis of the generated data . In proteomics and metabolomics,
[219]