Page 34 - Read Online
P. 34
Page 4 of 18 Fabbrini et al. Microbiome Res Rep 2023;2:25 https://dx.doi.org/10.20517/mrr.2023.25
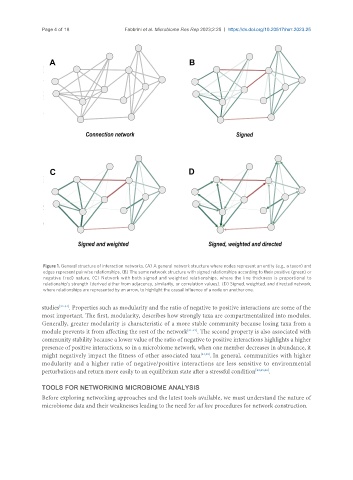
Figure 1. General structure of interaction networks. (A) A general network structure where nodes represent an entity (e.g., a taxon) and
edges represent pairwise relationships. (B) The same network structure with signed relationships according to their positive (green) or
negative (red) nature. (C) Network with both signed and weighted relationships, where the line thickness is proportional to
relationship’s strength (derived either from adjacency, similarity, or correlation values). (D) Signed, weighted, and directed network,
where relationships are represented by an arrow, to highlight the causal influence of a node on another one.
studies [32-34] . Properties such as modularity and the ratio of negative to positive interactions are some of the
most important. The first, modularity, describes how strongly taxa are compartmentalized into modules.
Generally, greater modularity is characteristic of a more stable community because losing taxa from a
module prevents it from affecting the rest of the network [31-43] . The second property is also associated with
community stability because a lower value of the ratio of negative to positive interactions highlights a higher
presence of positive interactions, so in a microbiome network, when one member decreases in abundance, it
might negatively impact the fitness of other associated taxa [34,35] . In general, communities with higher
modularity and a higher ratio of negative/positive interactions are less sensitive to environmental
perturbations and return more easily to an equilibrium state after a stressful condition [33,35,36] .
TOOLS FOR NETWORKING MICROBIOME ANALYSIS
Before exploring networking approaches and the latest tools available, we must understand the nature of
microbiome data and their weaknesses leading to the need for ad hoc procedures for network construction.