Page 106 - Read Online
P. 106
Page 6 of 12 Jones et al. Microbiome Res Rep 2024;3:24 https://dx.doi.org/10.20517/mrr.2023.78
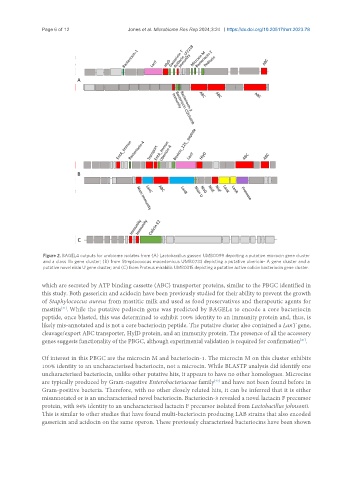
Figure 2. BAGEL4 outputs for urobiome isolates from (A) Lactobacillus gasseri UMB0099 depicting a putative microcin gene cluster
and a class IIb gene cluster; (B) from Streptococcus macedonicus UMB0733 depicting a putative ubericin- A gene cluster and a
putative novel nisin U gene cluster; and (C) from Proteus mirabilis UMB0315 depicting a putative active colicin bacteriocin gene cluster.
which are secreted by ATP binding cassette (ABC) transporter proteins, similar to the PBGC identified in
this study. Both gassericin and acidocin have been previously studied for their ability to prevent the growth
of Staphylococcus aureus from mastitic milk and used as food preservatives and therapeutic agents for
mastitis . While the putative pediocin gene was predicted by BAGEL4 to encode a core bacteriocin
[41]
peptide, once blasted, this was determined to exhibit 100% identity to an immunity protein and, thus, is
likely mis-annotated and is not a core bacteriocin peptide. The putative cluster also contained a LanT gene,
cleavage/export ABC transporter, HylD protein, and an immunity protein. The presence of all the accessory
genes suggests functionality of the PBGC, although experimental validation is required for confirmation .
[41]
Of interest in this PBGC are the microcin M and bacteriocin-1. The microcin M on this cluster exhibits
100% identity to an uncharacterised bacteriocin, not a microcin. While BLASTP analysis did identify one
uncharacterised bacteriocin, unlike other putative hits, it appears to have no other homologues. Microcins
are typically produced by Gram-negative Enterobacteriaceae family and have not been found before in
[42]
Gram-positive bacteria. Therefore, with no other closely related hits, it can be inferred that it is either
misannotated or is an uncharacterised novel bacteriocin. Bacteriocin-3 revealed a novel lactacin F precursor
protein, with 84% identity to an uncharacterised lactacin F precursor isolated from Lactobacillus johnsonii.
This is similar to other studies that have found multi-bacteriocin producing LAB strains that also encoded
gassericin and acidocin on the same operon. These previously characterised bacteriocins have been shown