Page 50 - Read Online
P. 50
Page 108 Marti et al. J Transl Genet Genom 2020;4:104-13 I http://dx.doi.org/10.20517/jtgg.2020.10
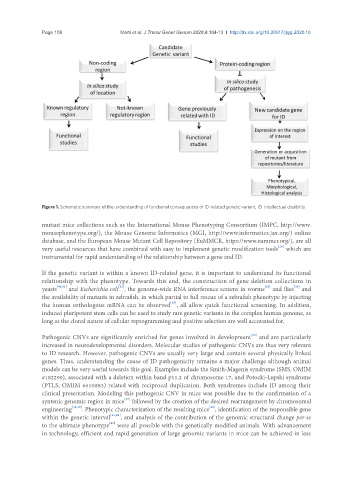
Figure 1. Schematic summary of the understanding of functional consequences of ID-related genetic variant. ID: intellectual disability
mutant mice collections such as the International Mouse Phenotyping Consortium (IMPC, http://www.
mousephenotype.org/), the Mouse Genome Informatics (MGI, http://www.informatics.jax.org/) online
database, and the European Mouse Mutant Cell Repository (EuMMCR, https://www.eummcr.org/), are all
[29]
very useful resources that have combined with easy to implement genetic modification tools which are
instrumental for rapid understanding of the relationship between a gene and ID.
If the genetic variant is within a known ID-related gene, it is important to understand its functional
relationship with the phenotype. Towards this end, the construction of gene deletion collections in
[33]
[34]
[32]
yeasts [30,31] and Escherichia coli , the genome-wide RNA interference screens in worms and flies and
the availability of mutants in zebrafish, in which partial to full rescue of a zebrafish phenotype by injecting
[35]
the human orthologous mRNA can be observed , all allow quick functional screening. In addition,
induced pluripotent stem cells can be used to study rare genetic variants in the complex human genome, as
long as the clonal nature of cellular reprogramming and positive selection are well accounted for.
[36]
Pathogenic CNVs are significantly enriched for genes involved in development and are particularly
increased in neurodevelopmental disorders. Molecular studies of pathogenic CNVs are thus very relevant
to ID research. However, pathogenic CNVs are usually very large and contain several physically linked
genes. Thus, understanding the cause of ID pathogenicity remains a major challenge although animal
models can be very useful towards this goal. Examples include the Smith-Magenis syndrome (SMS, OMIM
#182290), associated with a deletion within band p11.2 of chromosome 17, and Potocki-Lupski syndrome
(PTLS, OMIM #610883) related with reciprocal duplication. Both syndromes include ID among their
clinical presentation. Modeling this pathogenic CNV in mice was possible due to the confirmation of a
[37]
syntenic genomic region in mice followed by the creation of the desired rearrangement by chromosomal
engineering [38,39] . Phenotypic characterization of the resulting mice , identification of the responsible gene
[40]
within the genetic interval [41,42] , and analysis of the contribution of the genomic structural change per-se
to the ultimate phenotype were all possible with the genetically modified animals. With advancement
[43]
in technology, efficient and rapid generation of large genomic variants in mice can be achieved in less