Page 106 - Read Online
P. 106
Mohammadi et al. J Transl Genet Genom 2020;4:238-50 I https://doi.org/10.20517/jtgg.2020.29 Page 243
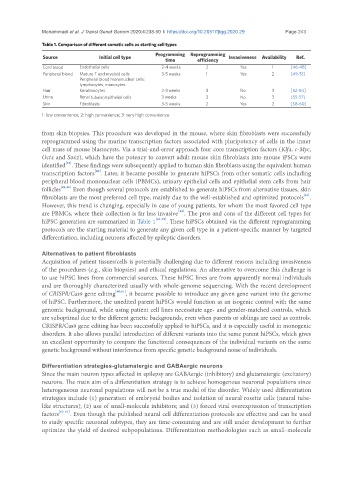
Table 1. Comparison of different somatic cells as starting cell types
Programming Reprogramming
Source Initial cell type Invasiveness Availability Ref.
time efficiency
Cord blood Endothelial cells 2-4 weeks 2 Yes 1 [46-48]
Peripheral blood Mature T and myeloid cells 3-5 weeks 1 Yes 2 [49-51]
Peripheral blood mononuclear cells:
lymphocytes, monocytes
Hair Keratinocytes 2-3 weeks 3 No 3 [52-54]
Urine Renal tubular/epithelial cells 3 weeks 3 No 3 [55-57]
Skin Fibroblasts 3-5 weeks 2 Yes 2 [58-60]
1: low convenience; 2: high convenience; 3: very high convenience
from skin biopsies. This procedure was developed in the mouse, where skin fibroblasts were successfully
reprogrammed using the murine transcription factors associated with pluripotency of cells in the inner
cell mass of mouse blastocysts. Via a trial-and-error approach four core transcription factors (Klf4, c-Myc,
Oct4 and Sox2), which have the potency to convert adult mouse skin fibroblasts into mouse iPSCs were
[39]
identified . These findings were subsequently applied to human skin fibroblasts using the equivalent human
[40]
transcription factors . Later, it became possible to generate hiPSCs from other somatic cells including
peripheral blood mononuclear cells (PBMCs), urinary epithelial cells and epithelial stem cells from hair
follicles [41,42] Even though several protocols are established to generate hiPSCs from alternative tissues, skin
[43]
fibroblasts are the most preferred cell type, mainly due to the well-established and optimized protocols .
However, this trend is changing, especially in case of young patients, for whom the most favored cell type
[44]
are PBMCs, where their collection is far less invasive . The pros and cons of the different cell types for
hiPSC generation are summarized in Table 1 [45-59] . These hiPSCs obtained via the different reprogramming
protocols are the starting material to generate any given cell type in a patient-specific manner by targeted
differentiation, including neurons affected by epileptic disorders.
Alternatives to patient fibroblasts
Acquisition of patient tissues/cells is potentially challenging due to different reasons including invasiveness
of the procedures (e.g., skin biopsies) and ethical regulations. An alternative to overcome this challenge is
to use hiPSC lines from commercial sources. These hiPSC lines are from apparently normal individuals
and are thoroughly characterized usually with whole-genome sequencing. With the recent development
of CRISPR/Cas9 gene editing [60,61] , it became possible to introduce any given gene variant into the genome
of hiPSC. Furthermore, the unedited parent hiPSCs would function as an isogenic control with the same
genomic background, while using patient cell lines necessitate age- and gender-matched controls, which
are suboptimal due to the different genetic backgrounds, even when parents or siblings are used as controls.
CRISPR/Cas9 gene editing has been successfully applied to hiPSCs, and it is especially useful in monogenic
disorders. It also allows parallel introduction of different variants into the same parent hiPSCs, which gives
an excellent opportunity to compare the functional consequences of the individual variants on the same
genetic background without interference from specific genetic background noise of individuals.
Differentiation strategies-glutamatergic and GABAergic neurons
Since the main neuron types affected in epilepsy are GABAergic (inhibitory) and glutamatergic (excitatory)
neurons. The main aim of a differentiation strategy is to achieve homogenous neuronal populations since
heterogeneous neuronal populations will not be a true model of the disorder. Widely used differentiation
strategies include (1) generation of embryoid bodies and isolation of neural rosette cells (neural tube-
like structures); (2) use of small-molecule inhibitors; and (3) forced viral overexpression of transcription
factors [62-64] . Even though the published neural cell differentiation protocols are effective and can be used
to study specific neuronal subtypes, they are time-consuming and are still under development to further
optimize the yield of desired subpopulations. Differentiation methodologies such as small-molecule