Page 67 - Read Online
P. 67
Page 4 of 15 Fraser. J Transl Genet Genom 2018;2:21. I https://doi.org/10.20517/jtgg.2018.27
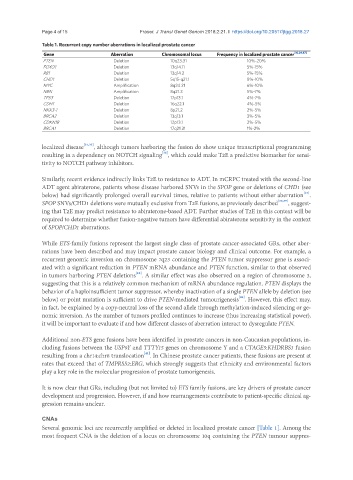
Table 1. Recurrent copy number aberrations in localized prostate cancer
Gene Aberration Chromosomal locus Frequency in localized prostate cancer [10,29,87]
PTEN Deletion 10q23.31 10%-20%
FOXO1 Deletion 13q14.11 5%-15%
RB1 Deletion 13q14.2 5%-15%
CHD1 Deletion 5q15-q21.1 8%-10%
MYC Amplification 8q24.21 6%-10%
NBN Amplification 8q21.3 5%-7%
TP53 Deletion 17p13.1 4%-7%
CDH1 Deletion 16q22.1 4%-5%
NKX3-1 Deletion 8p21.2 2%-5%
BRCA2 Deletion 13q13.1 3%-5%
CDKN1B Deletion 12p13.1 2%-5%
BRCA1 Deletion 17q21.31 1%-2%
localized disease [34,35] , although tumors harboring the fusion do show unique transcriptional programming
[36]
resulting in a dependency on NOTCH signaling , which could make T2E a predictive biomarker for sensi-
tivity to NOTCH pathway inhibitors.
Similarly, recent evidence indirectly links T2E to resistance to ADT. In mCRPC treated with the second-line
ADT agent abiraterone, patients whose disease harbored SNVs in the SPOP gene or deletions of CHD1 (see
[37]
below) had significantly prolonged overall survival times, relative to patients without either aberration .
SPOP SNVs/CHD1 deletions were mutually exclusive from T2E fusions, as previously described [38,39] , suggest-
ing that T2E may predict resistance to abiraterone-based ADT. Further studies of T2E in this context will be
required to determine whether fusion-negative tumors have differential abiraterone sensitivity in the context
of SPOP/CHD1 aberrations.
While ETS-family fusions represent the largest single class of prostate cancer-associated GRs, other aber-
rations have been described and may impact prostate cancer biology and clinical outcome. For example, a
recurrent genomic inversion on chromosome 7q23 containing the PTEN tumor suppressor gene is associ-
ated with a significant reduction in PTEN mRNA abundance and PTEN function, similar to that observed
[10]
in tumors harboring PTEN deletions . A similar effect was also observed on a region of chromosome 3,
suggesting that this is a relatively common mechanism of mRNA abundance regulation. PTEN displays the
behavior of a haploinsufficient tumor suppressor, whereby inactivation of a single PTEN allele by deletion (see
below) or point mutation is sufficient to drive PTEN-mediated tumourigenesis . However, this effect may,
[40]
in fact, be explained by a copy-neutral loss of the second allele through methylation-induced silencing or ge-
nomic inversion. As the number of tumors profiled continues to increase (thus increasing statistical power),
it will be important to evaluate if and how different classes of aberration interact to dysregulate PTEN.
Additional non-ETS gene fusions have been identified in prostate cancers in non-Caucasian populations, in-
cluding fusions between the USP9Y and TTTY15 genes on chromosome Y and a CTAGE5:KHDRBS3 fusion
[41]
resulting from a chr14:chr8 translocation . In Chinese prostate cancer patients, these fusions are present at
rates that exceed that of TMPRSS2:ERG, which strongly suggests that ethnicity and environmental factors
play a key role in the molecular progression of prostate tumorigenesis.
It is now clear that GRs, including (but not limited to) ETS family fusions, are key drivers of prostate cancer
development and progression. However, if and how rearrangements contribute to patient-specific clinical ag-
gression remains unclear.
CNAs
Several genomic loci are recurrently amplified or deleted in localized prostate cancer [Table 1]. Among the
most frequent CNA is the deletion of a locus on chromosome 10q containing the PTEN tumour suppres-