Page 64 - Read Online
P. 64
Page 4 of 18 Lee et al. J Cancer Metastasis Treat 2021;7:27 https://dx.doi.org/10.20517/2394-4722.2021.58
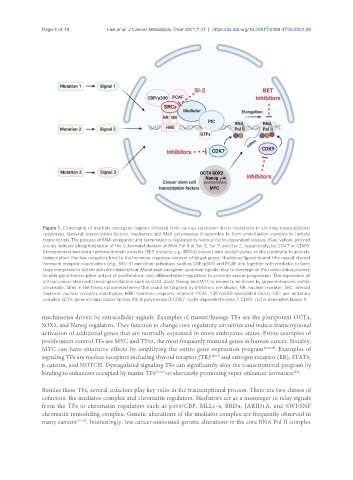
Figure 1. Converging of multiple oncogenic signals initiated from various upstream driver mutations in eliciting transcriptional
responses. General transcription factors, mediators and RNA polymerase II assemble to form preinitiation complex to initiate
transcription. The process of RNA elongation and termination is regulated by various cyclin-dependent kinases. Blue, yellow, and red
circles indicate phosphorylation of the C-terminal domain of RNA Pol II at Ser 5, Ser 7, and Ser 2, respectively, by CDK7 or CDK9.
Bromodomain and extra-terminal domain proteins (BET proteins; e.g., BRD4) interact with acetyl-lysines on the chromatin to activate
transcription. Nuclear receptors bind to the hormone response element of target genes. Hormone/ligand-bound HRs recruit steroid
hormone receptor coactivators (e.g., SRC-3) and other activators such as CBP/p300 and PCAF and together with mediator to form
large complexes to further activate transcription. Mutational oncogenic upstream signals relay to converge on the transcription process
to alter gene transcription output of proliferation- and differentiation-regulators to promote cancer progression. The expression of
critical cancer stem cell transcription factors such as Oct4, Sox2, Nanog and MYC is known to be driven by super-enhancers on the
chromatin. Sites in the transcription machinery that could be targeted by inhibitors are shown. NR: nuclear receptor; SRC: steroid
hormone nuclear receptor coactivator; HRE: hormone response element; PCAF: CBP/p300-associated factor; PIC: pre-initiation
complex; GTFs: general transcription factors; Pol II: polymerase II; CDK7: cyclin-dependent kinase 7; CDK9: cyclin-dependent kinase 9.
machineries driven by extracellular signals. Examples of master/lineage TFs are the pluripotent OCT4,
SOX2, and Nanog regulators. They function to change core regulatory circuitries and induce transcriptional
activation of additional genes that are normally expressed in more embryonic states. Prime examples of
proliferation control TFs are MYC and TP53, the most frequently mutated genes in human cancer. Notably,
MYC can have extensive effects by amplifying the entire gene expression program [4,47,48] . Examples of
signaling TFs are nuclear receptors including thyroid receptor (TR) [49,50] and estrogen receptor (ER), STAT3,
β-catenin, and NOTCH. Dysregulated signaling TFs can significantly alter the transcriptional program by
binding to enhancers occupied by master TFs [51,52] or aberrantly promoting super-enhancer formation .
[48]
Besides these TFs, several cofactors play key roles in the transcriptional process. There are two classes of
cofactors: the mediator complex and chromatin regulators. Mediators act as a messenger to relay signals
from the TFs to chromatin regulators such as p300/CBP, MLL1-4, BRD4, JARID1A, and SWI/SNF
chromatin remodeling complex. Genetic alterations of the mediator complex are frequently observed in
many cancers [53-55] . Interestingly, few cancer-associated genetic alterations in the core RNA Pol II complex