Page 72 - Read Online
P. 72
Malentacchi et al. J Cancer Metastasis Treat 2020;6:34 I http://dx.doi.org/10.20517/2394-4722.2020.34 Page 5 of 18
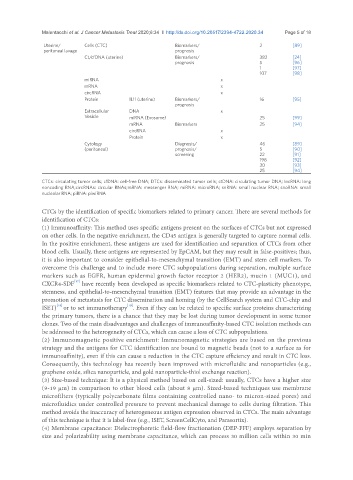
Uterine/ Cells (CTC) Biomarkers/ 2 [89]
peritoneal lavage prognosis
Ct/cfDNA (uterine) Biomarkers/ 382 [24]
prognosis 5 [96]
1 [97]
107 [98]
miRNA x
mRNA x
circRNA x
Protein IL11 (uterine) Biomarkers/ 16 [95]
prognosis
Extracellular DNA x
Vesicle miRNA (Exosome) 25 [99]
mRNA Biomarkers 25 [94]
circRNA x
Protein x
Cytology Diagnosis/ 46 [89]
(peritoneal) prognosis/ 5 [90]
screening 22 [91]
198 [92]
30 [93]
25 [94]
CTCs: circulating tumor cells; cfDNA: cell-free DNA; DTCs: disseminated tumor cells; ctDNA: circulating tumor DNA; lncRNA: long
noncoding RNA;circRNAs: circular RNAs;mRNA: messenger RNA; miRNA: microRNA; snRNA: small nuclear RNA; snoRNA: small
nucleolar RNA; piRNA: piwiRNA
CTCs by the identification of specific biomarkers related to primary cancer. There are several methods for
identification of CTCs:
(1) Immunoaffinity: This method uses specific antigens present on the surfaces of CTCs but not expressed
on other cells. In the negative enrichment, the CD45 antigen is generally targeted to capture normal cells.
In the positive enrichment, these antigens are used for identification and separation of CTCs from other
blood cells. Usually, these antigens are represented by EpCAM, but they may result in false-positives; thus,
it is also important to consider epithelial-to-mesenchymal transition (EMT) and stem cell markers. To
overcome this challenge and to include more CTC subpopulations during separation, multiple surface
markers such as EGFR, human epidermal growth factor receptor 2 (HER2), mucin 1 (MUC1), and
[17]
CXCR4-SDF have recently been developed as specific biomarkers related to CTC-plasticity phenotype,
stemness, and epithelial-to-mesenchymal transition (EMT) features that may provide an advantage in the
promotion of metastasis for CTC dissemination and homing (by the CellSearch system and CTC-chip and
[19]
ISET) or to set immunotherapy . Even if they can be related to specific surface proteins characterizing
[18]
the primary tumors, there is a chance that they may be lost during tumor development in some tumor
clones. Two of the main disadvantages and challenges of immunoaffinity-based CTC isolation methods can
be addressed to the heterogeneity of CTCs, which can cause a loss of CTC subpopulations.
(2) Immunomagnetic positive enrichment: Immunomagnetic strategies are based on the previous
strategy and the antigens for CTC identification are bound to magnetic beads (not to a surface as for
immunoaffinity), even if this can cause a reduction in the CTC capture efficiency and result in CTC loss.
Consequently, this technology has recently been improved with microfluidic and nanoparticles (e.g.,
graphene oxide, silica nanoparticle, and gold nanoparticle-thiol exchange reaction).
(3) Size-based technique: It is a physical method based on cell-sized: usually, CTCs have a higher size
(9-19 μm) in comparison to other blood cells (about 8 μm). Sized-based techniques use membrane
microfilters (typically polycarbonate films containing controlled nano- to micron-sized pores) and
microfluidics under controlled pressure to prevent mechanical damage to cells during filtration. This
method avoids the inaccuracy of heterogeneous antigen expression observed in CTCs. The main advantage
of this technique is that it is label-free (e.g., ISET, ScreenCellCyto, and Parasortix).
(4) Membrane capacitance: Dielectrophoretic field-flow fractionation (DEP-FFF) employs separation by
size and polarizability using membrane capacitance, which can process 30 million cells within 30 min