Page 71 - Read Online
P. 71
Miliotis et al. J Cancer Metastasis Treat 2020;6:13 I http://dx.doi.org/10.20517/2394-4722.2020.12 Page 5 of 15
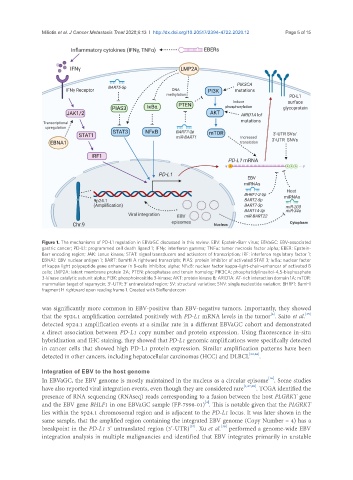
Figure 1. The mechanisms of PD-L1 regulation in EBVaGC discussed in this review. EBV: Epstein-Barr virus; EBVaGC: EBV-associated
gastric cancer; PD-L1: programmed cell death ligand 1; IFNγ: interferon gamma; TNFα: tumor necrosis factor alpha; EBER: Epstein-
Barr encoding region; JAK: Janus kinase; STAT: signal transducers and activators of transcription; IRF: interferon regulatory factor 1;
EBNA1: EBV nuclear antigen 1; BART: BamHI A rightward transcripts; PIAS: protein inhibitor of activated STAT 3; IκBα: nuclear factor
of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha; NFκB: nuclear factor kappa-light-chain-enhancer of activated B
cells; LMP2A: latent membrane protein 2A; PTEN: phosphatase and tensin homolog; PIK3CA: phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit alpha; PI3K: phosphoinositide 3-kinase; AKT: protein kinase B; ARID1A: AT-rich interaction domain 1A; mTOR:
mammalian target of rapamycin; 3′-UTR: 3′ untranslated region; SV: structural variation; SNV: single nucleotide variation; BHRF1: BamHI
fragment H rightward open reading frame 1. Created with BioRender.com
was significantly more common in EBV-positive than EBV-negative tumors. Importantly, they showed
[4]
[35]
that the 9p24.1 amplification correlated positively with PD-L1 mRNA levels in the tumor . Saito et al.
detected 9p24.1 amplification events at a similar rate in a different EBVaGC cohort and demonstrated
a direct association between PD-L1 copy number and protein expression. Using fluorescence in-situ
hybridization and IHC staining, they showed that PD-L1 genomic amplifications were specifically detected
in cancer cells that showed high PD-L1 protein expression. Similar amplification patterns have been
detected in other cancers, including hepatocellular carcinomas (HCC) and DLBCL [23,36] .
Integration of EBV to the host genome
[16]
In EBVaGC, the EBV genome is mostly maintained in the nucleus as a circular episome . Some studies
have also reported viral integration events, even though they are considered rare [4,37,38] . TCGA identified the
presence of RNA sequencing (RNAseq) reads corresponding to a fusion between the host PLGRKT gene
[4]
and the EBV gene BHLF1 in one EBVaGC sample (FP-7998-01) . This is notable given that the PLGRKT
lies within the 9p24.1 chromosomal region and is adjacent to the PD-L1 locus. It was later shown in the
same sample, that the amplified region containing the integrated EBV genome (Copy Number = 4) has a
[38]
[37]
breakpoint in the PD-L1 3’ untranslated region (3’-UTR) . Xu et al. performed a genome-wide EBV
integration analysis in multiple malignancies and identified that EBV integrates primarily in unstable