Page 93 - Read Online
P. 93
Page 8 of 11 Kaufman et al. J Cancer Metastasis Treat 2019;5:73 I http://dx.doi.org/10.20517/2394-4722.2019.19
3D models
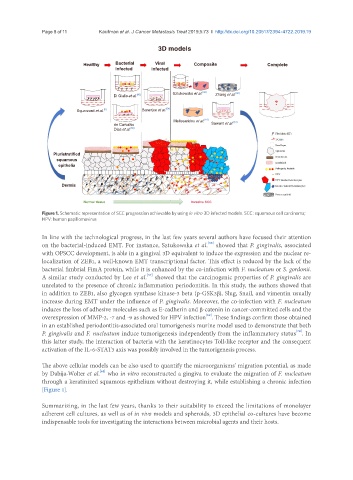
Healthy Bacterial Viral Composite Complete
infected infected
Sztukowska et al. [38] [60]
Di Giulio et al. [50] Zhang et al.
Squarzanti et al. [5] Banerjee et al. [54]
Melissaridou et al. [16] [61]
de Carvalho Sawant et al.
Dias et al. [59]
Pluristratified
squamous
epithelia
Dermis
Normal tissue Invasive SCC
Figure 1. Schematic representation of SCC progression achievable by using in vitro 3D infected models. SCC: squamous cell carcinoma;
HPV: human papillomavirus
In line with the technological progress, in the last few years several authors have focused their attention
[38]
on the bacterial-induced EMT. For instance, Sztukowska et al. showed that P. gingivalis, associated
with OPSCC development, is able in a gingival 3D equivalent to induce the expression and the nuclear re-
localization of ZEB1, a well-known EMT transcriptional factor. This effect is reduced by the lack of the
bacterial fimbrial FimA protein, while it is enhanced by the co-infection with F. nucleatum or S. gordonii.
[37]
A similar study conducted by Lee et al. showed that the carcinogenic properties of P. gingivalis are
unrelated to the presence of chronic inflammation periodontitis. In this study, the authors showed that
in addition to ZEB1, also glycogen synthase kinase-3 beta (p-GSK3β), Slug, Snail, and vimentin usually
increase during EMT under the influence of P. gingivalis. Moreover, the co-infection with F. nucleatum
induces the loss of adhesive molecules such as E-cadherin and β-catenin in cancer-committed cells and the
overexpression of MMP-2, -7 and -9 as showed for HPV infection . These findings confirm those obtained
[69]
in an established periodontitis-associated oral tumorigenesis murine model used to demonstrate that both
P. gingivalis and F. nucleatum induce tumorigenesis independently from the inflammatory status . In
[70]
this latter study, the interaction of bacteria with the keratinocytes Toll-like receptor and the consequent
activation of the IL-6-STAT3 axis was possibly involved in the tumorigenesis process.
The above cellular models can be also used to quantify the microorganisms’ migration potential, as made
[62]
by Dabija-Wolter et al. who in vitro reconstructed a gingiva to evaluate the migration of F. nucleatum
through a keratinized squamous epithelium without destroying it, while establishing a chronic infection
[Figure 1].
Summarizing, in the last few years, thanks to their suitability to exceed the limitations of monolayer
adherent cell cultures, as well as of in vivo models and spheroids, 3D epithelial co-cultures have become
indispensable tools for investigating the interactions between microbial agents and their hosts.