Page 82 - Read Online
P. 82
Moia et al. J Cancer Metastasis Treat 2019;5:67 I http://dx.doi.org/10.20517/2394-4722.2019.020 Page 5 of 8
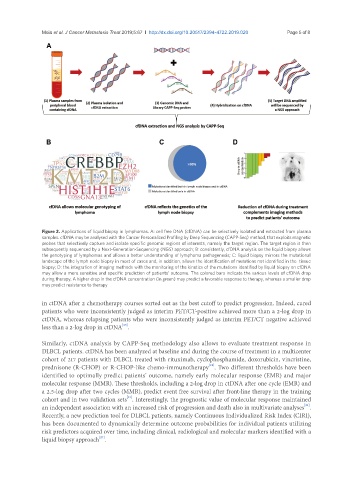
Figure 2. Applications of liquid biopsy in lymphomas. A: cell free DNA (cfDNA) can be selectively isolated and extracted from plasma
samples. cfDNA may be analysed with the Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) method, that exploits magnetic
probes that selectively capture and isolate specific genomic regions of interests, namely the target region. The target region is then
subsequently sequenced by a Next-Generation-Sequencing (NGS) approach; B: consistently, cfDNA analysis on the liquid biopsy allows
the genotyping of lymphomas and allows a better understanding of lymphoma pathogenesis; C: liquid biopsy mirrors the mutational
landscape of the lymph node biopsy in most of cases and, in addition, allows the identification of mutations not identified in the tissue
biopsy; D: the integration of imaging methods with the monitoring of the kinetics of the mutations identified by liquid biopsy on cfDNA
may allow a more sensitive and specific prediction of patients’ outcome. The colored bars indicate the various levels of cfDNA drop
during therapy. A higher drop in the cfDNA concentration (in green) may predict a favorable response to therapy, whereas a smaller drop
may predict resistance to therapy
in ctDNA after 2 chemotherapy courses sorted out as the best cutoff to predict progression. Indeed, cured
patients who were inconsistently judged as interim PET/CT-positive achieved more than a 2-log drop in
ctDNA, whereas relapsing patients who were inconsistently judged as interim PET/CT negative achieved
[23]
less than a 2-log drop in ctDNA .
Similarly, ctDNA analysis by CAPP-Seq methodology also allows to evaluate treatment response in
DLBCL patients. ctDNA has been analyzed at baseline and during the course of treatment in a multicenter
cohort of 217 patients with DLBCL treated with rituximab, cyclophosphamide, doxorubicin, vincristine,
[21]
prednisone (R-CHOP) or R-CHOP-like chemo-immunotherapy . Two different thresholds have been
identified to optimally predict patients’ outcome, namely early molecular response (EMR) and major
molecular response (MMR). These thresholds, including a 2-log drop in ctDNA after one cycle (EMR) and
a 2.5-log drop after two cycles (MMR), predict event free survival after front-line therapy in the training
[21]
cohort and in two validation sets . Interestingly, the prognostic value of molecular response maintained
an independent association with an increased risk of progression and death also in multivariate analyses .
[21]
Recently, a new prediction tool for DLBCL patients, namely Continuous Individualized Risk Index (CIRI),
has been documented to dynamically determine outcome probabilities for individual patients utilizing
risk predictors acquired over time, including clinical, radiological and molecular markers identified with a
[27]
liquid biopsy approach .