Page 363 - Read Online
P. 363
Page 8 of 16 Anstine et al. J Cancer Metastasis Treat 2019;5:50 I http://dx.doi.org/10.20517/2394-4722.2019.24
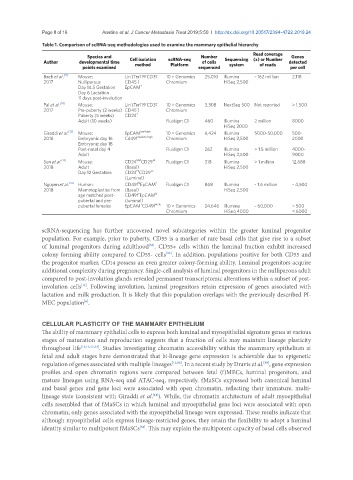
Table 1. Comparison of scRNA-seq methodologies used to examine the mammary epithelial hierarchy
Species and Number Read coverage Genes
Author developmental time Cell isolation scRNA-seq of cells Sequencing (x) or Number detected
points examined method Platform sequenced system of reads per cell
-
-
Bach et al. [11] Mouse: Lin (Ter119 CD31 - 10 × Genomics 25,010 Illumina ~ 162 million 2,118
-
2017 Nulliparous CD45 ) Chromium HiSeq 2,500
Day 14.5 Gestation EpCAM +
Day 6 Lactation
11 days post-involution
-
-
Pal et al. [53] Mouse: Lin (Ter119 CD31 - 10 × Genomics 3,308 NextSeq 500 Not reported > 1,500
-
2017 Pre-puberty (2 weeks) CD45 ) Chromium
Puberty (5 weeks) CD24 +
Adult (10 weeks) Fluidigm C1 460 Illumina 2 million 8000
HiSeq 2000
[12]
Giraddi et al. Mouse: EpCAM low-high 10 × Genomics 6,424 Illumina 5000-50,000 500-
2018 Embryonic day 16 Cd49f medium-high Chromium HiSeq 2,500 2000
Embryonic day 18
Post-natal day 4 Fluidigm C1 262 Illumina > 1.5 million 4000-
Adult HiSeq 2,500 9000
Hi
Sun et al. Mouse: CD24 CD29 Fluidigm C1 318 Illumina > 1 million 12,688
[13]
Mid
2018 Adult (Basal) HiSeq 2,500
Day 12 Gestation CD24 CD29
Lo
Hi
(Luminal)
hi
Nguyen et al. [14] Human: CD49f EpCAM + Fluidigm C1 868 Illumina ~ 1.6 million ~ 4,500
2018 Mammoplasties from (Basal) HiSeq 2,500
+
hi
age matched post- CD49f EpCAM
pubertal and pre- (luminal)
+
pubertal females EpCAM CD49f hi/lo 10 × Genomics 24,646 Illumina ~ 60,000 > 500
Chromium HiSeq 4000 < 6000
scRNA-sequencing has further uncovered novel subcategories within the greater luminal progenitor
population. For example, prior to puberty, CD55 is a marker of rare basal cells that give rise to a subset
of luminal progenitors during adulthood . CD55+ cells within the luminal fraction exhibit increased
[53]
colony forming ability compared to CD55- cells . In addition, populations positive for both CD55 and
[53]
the progenitor marker, CD14 possess an even greater colony-forming ability. Luminal progenitors acquire
additional complexity during pregnancy. Single-cell analysis of luminal progenitors in the nulliparous adult
compared to post-involution glands revealed permanent transcriptomic alterations within a subset of post-
involution cells . Following involution, luminal progenitors retain expression of genes associated with
[11]
lactation and milk production. It is likely that this population overlaps with the previously described PI-
MEC population .
[6]
CELLULAR PLASTICITY OF THE MAMMARY EPITHELIUM
The ability of mammary epithelial cells to express both luminal and myoepithelial signature genes at various
stages of maturation and reproduction suggests that a fraction of cells may maintain lineage plasticity
throughout life [12,14,52,53] . Studies investigating chromatin accessibility within the mammary epithelium at
fetal and adult stages have demonstrated that bi-lineage gene expression is achievable due to epigenetic
regulation of genes associated with multiple lineages [12,58] . In a recent study by Dravis et al. , gene expression
[58]
profiles and open chromatin regions were compared between fetal (f)MECs, luminal progenitors, and
mature lineages using RNA-seq and ATAC-seq, respectively. fMaSCs expressed both canonical luminal
and basal genes and gene loci were associated with open chromatin, reflecting their immature, multi-
lineage state (consistent with Giraddi et al. ). While, the chromatin architecture of adult myoepithelial
[12]
cells resembled that of fMaSCs in which luminal and myoepithelial gene loci were associated with open
chromatin; only genes associated with the myoepithelial lineage were expressed. These results indicate that
although myoepithelial cells express lineage-restricted genes, they retain the flexibility to adopt a luminal
identity similar to multipotent fMaSCs . This may explain the multipotent capacity of basal cells observed
[58]