Page 999 - Read Online
P. 999
Page 2 of 10 Kim et al. Hepatoma Res 2020;6:85 I http://dx.doi.org/10.20517/2394-5079.2020.96
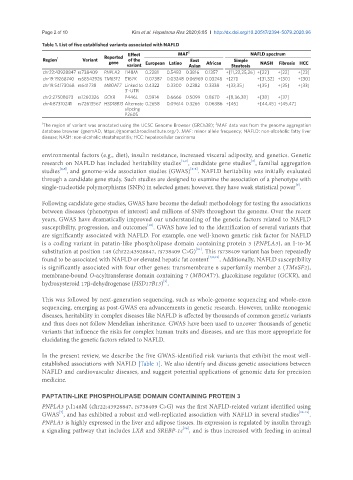
Table 1. List of five established variants associated with NAFLD 2
Region 1 Variant Reported Effect MAF East Simple NAFLD spectrum
of the
gene
variant European Latino Asian African Steatosis NASH Fibrosis HCC
chr22:43928847 rs738409 PNPLA3 I148M 0.2281 0.5493 0.3816 0.1357 +[11,22,25,26] +[22] +[22] +[23]
chr19:19268740 rs58542926 TM6SF2 E167K 0.07387 0.03248 0.06969 0.03248 +[27] +[31,32] +[30] +[30]
chr19:54173068 rs641738 MBOAT7 Linked to 0.4322 0.3300 0.2382 0.3338 +[33,35] +[35] +[35] +[33]
3’-UTR
chr2:27508073 rs1260326 GCKR P446L 0.5914 0.6666 0.5099 0.8670 +[8,36,38] +[38] +[37]
chr4:87310241 rs72613567 HSD18B13 Alternate 0.2658 0.09614 0.3266 0.06386 +[45] +[44,45] +[45,47]
slipcing
P260S
2
1 The region of variant was annotated using the UCSC Genome Browser (GRCh38); MAF data was from the genome aggregation
database browser (gnomAD, https://gnomad.broadinstitute.org/). MAF: minor allele frequency; NAFLD: non-alcoholic fatty liver
disease; NASH: non-alcoholic steatohepatitis; HCC: hepatocellular carcinoma
environmental factors (e.g., diet), insulin resistance, increased visceral adiposity, and genetics. Genetic
[3]
[1,2]
research on NAFLD has included heritability studies , candidate gene studies , familial aggregation
[6-8]
[4,5]
studies , and genome-wide association studies (GWAS) . NAFLD heritability was initially evaluated
through a candidate gene study. Such studies are designed to examine the association of a phenotype with
single-nucleotide polymorphisms (SNPs) in selected genes; however, they have weak statistical power .
[9]
Following candidate gene studies, GWAS have become the default methodology for testing the associations
between diseases (phenotypes of interest) and millions of SNPs throughout the genome. Over the recent
years, GWAS have dramatically improved our understanding of the genetic factors related to NAFLD
[10]
susceptibility, progression, and outcomes . GWAS have led to the identification of several variants that
are significantly associated with NAFLD. For example, one well-known genetic risk factor for NAFLD
is a coding variant in patatin-like phospholipase domain containing protein 3 (PNPLA3), an I-to-M
substitution at position 148 (chr22:43928847, rs738409 C>G) . This rs738409 variant has been repeatedly
[11]
found to be associated with NAFLD or elevated hepatic fat content [7,8,12] . Additionally, NAFLD susceptibility
is significantly associated with four other genes: transmembrane 6 superfamily member 2 (TM6SF2),
membrane-bound O-acyltransferase domain containing 7 (MBOAT7), glucokinase regulator (GCKR), and
hydroxysteroid 17β-dehydrogenase (HSD17B13) .
[9]
This was followed by next-generation sequencing, such as whole-genome sequencing and whole-exon
sequencing, emerging as post-GWAS era advancements in genetic research. However, unlike monogenic
diseases, heritability in complex diseases like NAFLD is affected by thousands of common genetic variants
and thus does not follow Mendelian inheritance. GWAS have been used to uncover thousands of genetic
variants that influence the risks for complex human traits and diseases, and are thus more appropriate for
elucidating the genetic factors related to NAFLD.
In the present review, we describe the five GWAS-identified risk variants that exhibit the most well-
established associations with NAFLD [Table 1]. We also identify and discuss genetic associations between
NAFLD and cardiovascular diseases, and suggest potential applications of genomic data for precision
medicine.
PAPTATIN-LIKE PHOSPHOLIPASE DOMAIN CONTAINING PROTEIN 3
PNPLA3 p.I148M (chr22:43928847, rs738409 C>G) was the first NAFLD-related variant identified using
[7]
GWAS , and has exhibited a robust and well-replicated association with NAFLD in several studies [13-15] .
PNPLA3 is highly expressed in the liver and adipose tissues. Its expression is regulated by insulin through
[16]
a signaling pathway that includes LXR and SREBP-1c , and is thus increased with feeding in animal