Page 99 - Read Online
P. 99
Ma et al. Hepatoma Res 2019;5:8 I http://dx.doi.org/10.20517/2394-5079.2018.104 Page 3 of 12
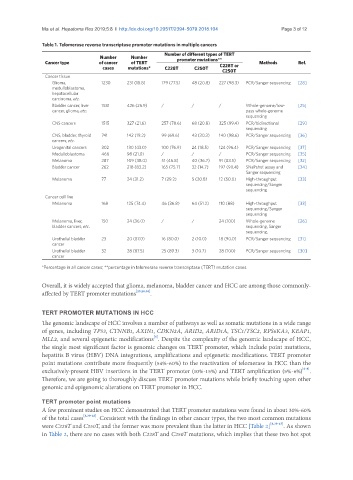
Table 1. Telomerase reverse transcriptase promoter mutations in multiple cancers
Number of different types of TERT
Number Number promoter mutations**
Cancer type of cancer of TERT Methods Ref.
cases mutations* C228T C250T C228T or
C250T
Cancer tissue
Glioma, 1230 231 (18.8) 179 (77.5) 48 (20.8) 227 (98.3) PCR/Sanger sequencing [28]
medulloblastoma,
hepatocellular
carcinoma, etc.
Bladder cancer, liver 1581 426 (26.9) / / / Whole-genome/low- [25]
cancer, glioma, etc. pass whole-genome
sequencing
CNS cancers 1515 327 (21.6) 257 (78.6) 68 (20.8) 325 (99.4) PCR/bidirectional [29]
sequencing
CNS, bladder, thyroid 741 142 (19.2) 99 (69.6) 43 (30.3) 140 (98.6) PCR/Sanger sequencing [36]
cancers, etc.
Urogenital cancers 302 130 (43.0) 100 (76.9) 24 (18.5) 124 (96.4) PCR/Sanger sequencing [37]
Medulloblastoma 466 98 (21.0) / / / PCR/Sanger sequencing [35]
Melanoma 287 109 (38.0) 51 (46.8) 40 (36.7) 91 (83.5) PCR/Sanger sequencing [32]
Bladder cancer 262 218 (83.2) 165 (75.7) 32 (14.7) 197 (90.4) SNaPshot assay and [34]
Sanger sequencing
Melanoma 77 24 (31.2) 7 (29.2) 5 (20.8) 12 (50.0) High-throughput [33]
sequencing/Sanger
sequencing
Cancer cell line
Melanoma 168 125 (74.4) 46 (36.8) 64 (51.2) 110 (88) High-throughput [33]
sequencing/Sanger
sequencing
Melanoma, liver, 150 24 (36.0) / / 24 (100) Whole-genome [26]
bladder cancers, etc. sequencing, Sanger
sequencing,
Urothelial bladder 23 20 (87.0) 16 (80.0) 2 (10.0) 18 (90.0) PCR/Sanger sequencing [31]
cancer
Urothelial bladder 32 28 (87.5) 25 (89.3) 3 (10.7) 28 (100) PCR/Sanger sequencing [30]
cancer
*Percentage in all cancer cases; **percentage in telomerase reverse transcriptase (TERT) mutation cases
Overall, it is widely accepted that glioma, melanoma, bladder cancer and HCC are among those commonly-
affected by TERT promoter mutations [25,28,38] .
TERT PROMOTER MUTATIONS IN HCC
The genomic landscape of HCC involves a number of pathways as well as somatic mutations in a wide range
of genes, including TP53, CTNNB1, AXIN1, CDKN2A, ARID2, ARID1A, TSC1/TSC2, RPS6KA3, KEAP1,
[6]
MLL2, and several epigenetic modifications . Despite the complexity of the genomic landscape of HCC,
the single most significant factor is genomic changes on TERT promoter, which include point mutations,
hepatitis B virus (HBV) DNA integrations, amplifications and epigenetic modifications. TERT promoter
point mutations contribute more frequently (54%-60%) to the reactivation of telomerase in HCC than the
[6-8]
exclusively-present HBV insertions in the TERT promoter (10%-15%) and TERT amplification (5%-6%) .
Therefore, we are going to thoroughly discuss TERT promoter mutations while briefly touching upon other
genomic and epigenomic alterations on TERT promoter in HCC.
TERT promoter point mutations
A few prominent studies on HCC demonstrated that TERT promoter mutations were found in about 30%-60%
of the total cases [8,39-49] . Consistent with the findings in other cancer types, the two most common mutations
were C228T and C250T, and the former was more prevalent than the latter in HCC [Table 2] [8,39-47] . As shown
in Table 2, there are no cases with both C228T and C250T mutations, which implies that these two hot spot