Page 49 - Read Online
P. 49
Liu et al. Hepatoma Res 2020;6:7 I http://dx.doi.org/10.20517/2394-5079.2019.39 Page 3 of 16
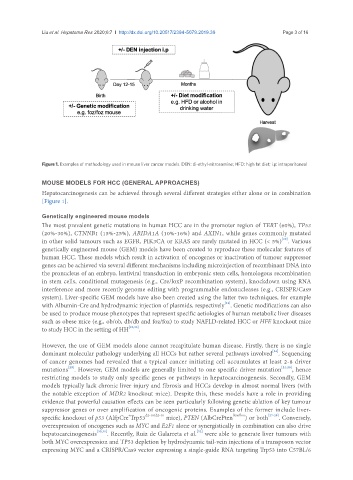
Figure 1. Examples of methodology used in mouse liver cancer models. DEN: di-ethyl-nitrosamine; HFD: high fat diet; i.p: intraperitoneal
MOUSE MODELS FOR HCC (GENERAL APPROACHES)
Hepatocarcinogenesis can be achieved through several different strategies either alone or in combination
[Figure 1].
Genetically engineered mouse models
The most prevalent genetic mutations in human HCC are in the promoter region of TERT (60%), TP53
(20%-30%), CTNNB1 (15%-25%), ARIDA1A (10%-16%) and AXIN1, while genes commonly mutated
[20]
in other solid tumours such as EGFR, PIK3CA or KRAS are rarely mutated in HCC (< 5%) . Various
genetically engineered mouse (GEM) models have been created to reproduce these molecular features of
human HCC. These models which result in activation of oncogenes or inactivation of tumour suppressor
genes can be achieved via several different mechanisms including microinjection of recombinant DNA into
the pronucleus of an embryo, lentiviral transduction in embryonic stem cells, homologous recombination
in stem cells, conditional mutagenesis (e.g., Cre/loxP recombination system), knockdown using RNA
interference and more recently genome editing with programmable endonucleases (e.g., CRISPR/Cas9
system). Liver-specific GEM models have also been created using the latter two techniques, for example
[21]
with Albumin-Cre and hydrodynamic injection of plasmids, respectively . Genetic modifications can also
be used to produce mouse phenotypes that represent specific aetiologies of human metabolic liver diseases
such as obese mice (e.g., ob/ob, db/db and foz/foz) to study NAFLD-related HCC or HFE knockout mice
to study HCC in the setting of HH [22,23] .
However, the use of GEM models alone cannot recapitulate human disease. Firstly, there is no single
dominant molecular pathology underlying all HCCs but rather several pathways involved . Sequencing
[24]
of cancer genomes had revealed that a typical cancer initiating cell accumulates at least 2-8 driver
[25]
mutations . However, GEM models are generally limited to one specific driver mutation [21,26] , hence
restricting models to study only specific genes or pathways in hepatocarcinogenesis. Secondly, GEM
models typically lack chronic liver injury and fibrosis and HCCs develop in almost normal livers (with
the notable exception of MDR2 knockout mice). Despite this, these models have a role in providing
evidence that powerful causation effects can be seen particularly following genetic ablation of key tumour
suppressor genes or over amplification of oncogenic proteins. Examples of the former include liver-
specific knockout of p53 (AlfpCre Trp53 Δ2-10/Δ2-10 mice), PTEN (AlbCrePten flox/flox ) or both [27-29] . Conversely,
+
overexpression of oncogenes such as MYC and E2F1 alone or synergistically in combination can also drive
hepatocarcinogenesis [30,31] . Recently, Ruiz de Galarreta et al. were able to generate liver tumours with
[32]
both MYC overexpression and TP53 depletion by hydrodynamic tail-vein injections of a transposon vector
expressing MYC and a CRISPR/Cas9 vector expressing a single-guide RNA targeting Trp53 into C57BL/6